Structural variants in the Epb41l4a locus: TAD disruption and Nrep gene misregulation as hypothetical drivers of neurodevelopmental outcomes
- PMID: 38438377
- PMCID: PMC10912600
- DOI: 10.1038/s41598-024-52545-y
Structural variants in the Epb41l4a locus: TAD disruption and Nrep gene misregulation as hypothetical drivers of neurodevelopmental outcomes
Abstract
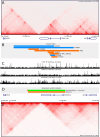
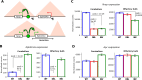
Structural variations are a pervasive feature of human genomes, and there is growing recognition of their role in disease development through their impact on spatial chromatin architecture. This understanding has led us to investigate the clinical significance of CNVs in noncoding regions that influence TAD structures. In this study, we focused on the Epb41l4a locus, which contains a highly conserved TAD boundary present in both human chromosome 5 and mouse chromosome 18, and its association with neurodevelopmental phenotypes. Analysis of human data from the DECIPHER database indicates that CNVs within this locus, including both deletions and duplications, are often observed alongside neurological abnormalities, such as dyslexia and intellectual disability, although there is not enough evidence of a direct correlation or causative relationship. To investigate these possible associations, we generated mouse models with deletion and inversion mutations at this locus and carried out RNA-seq analysis to elucidate gene expression changes. We found that modifications in the Epb41l4a TAD boundary led to dysregulation of the Nrep gene, which plays a crucial role in nervous system development. These findings underscore the potential pathogenicity of these CNVs and highlight the crucial role of spatial genome architecture in gene expression regulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The effects of common structural variants on 3D chromatin structure.BMC Genomics. 2020 Jan 30;21(1):95. doi: 10.1186/s12864-020-6516-1. BMC Genomics. 2020. PMID: 32000688 Free PMC article.
-
Formation of new chromatin domains determines pathogenicity of genomic duplications.Nature. 2016 Oct 13;538(7624):265-269. doi: 10.1038/nature19800. Epub 2016 Oct 5. Nature. 2016. PMID: 27706140
-
Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions.Cell. 2015 May 21;161(5):1012-1025. doi: 10.1016/j.cell.2015.04.004. Epub 2015 May 7. Cell. 2015. PMID: 25959774 Free PMC article.
-
16p11.2 Copy Number Variations and Neurodevelopmental Disorders.Trends Neurosci. 2020 Nov;43(11):886-901. doi: 10.1016/j.tins.2020.09.001. Epub 2020 Sep 28. Trends Neurosci. 2020. PMID: 32993859 Free PMC article. Review.
-
TAD disruption as oncogenic driver.Curr Opin Genet Dev. 2016 Feb;36:34-40. doi: 10.1016/j.gde.2016.03.008. Epub 2016 Apr 22. Curr Opin Genet Dev. 2016. PMID: 27111891 Free PMC article. Review.
Cited by
-
Loop Extrusion Machinery Impairments in Models and Disease.Cells. 2024 Nov 17;13(22):1896. doi: 10.3390/cells13221896. Cells. 2024. PMID: 39594644 Free PMC article. Review.
-
Structural Variants: Mechanisms, Mapping, and Interpretation in Human Genetics.Genes (Basel). 2025 Jul 29;16(8):905. doi: 10.3390/genes16080905. Genes (Basel). 2025. PMID: 40869953 Free PMC article. Review.
-
Structural variants in the 3D genome as drivers of disease.Nat Rev Genet. 2025 Jun 30. doi: 10.1038/s41576-025-00862-x. Online ahead of print. Nat Rev Genet. 2025. PMID: 40588575 Review.
-
Direction and modality of transcription changes caused by TAD boundary disruption in Slc29a3/Unc5b locus depends on tissue-specific epigenetic context.Epigenetics Chromatin. 2025 Aug 12;18(1):55. doi: 10.1186/s13072-025-00618-1. Epigenetics Chromatin. 2025. PMID: 40796890 Free PMC article.
References
-
- Wang S, et al. The 3D genome and its impacts on human health and disease. Life Med. 2023;2:lnad012. doi: 10.1093/lifemedi/lnad012. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

