The minimum energy required to build a cell
- PMID: 38438463
- PMCID: PMC11306549
- DOI: 10.1038/s41598-024-54303-6
The minimum energy required to build a cell
Abstract
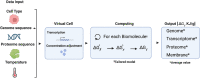
Understanding the energy requirements for cell synthesis accurately and comprehensively has been a longstanding challenge. We introduce a computational model that estimates the minimum energy necessary to build any cell from its constituent parts. This method combines omics and internal cell compositions from various sources to calculate the Gibbs Free Energy of biosynthesis independently of specific metabolic pathways. Our public tool, Synercell, can be used with other models for minumum species-specific energy estimations in any well-sequenced species. The energy for synthesising the genome, transcriptome, proteome, and lipid bilayer of four cell types: Escherichia coli, Saccharomyces cerevisiae, an average mammalian cell and JCVI-syn3A were estimated. Their modelled minimum synthesis energies at 298 K were J/cell, J/cell, J/cell and respectively. Gram-for-gram synthesis of lipid bilayers requires the most energy, followed by the proteome, genome, and transcriptome. The average per gram cost of biomass synthesis is in the 300s of J/g for all four cells. Implications for the generalisability of cell construction and applications to biogeosciences, cellular biology, biotechnology, and astrobiology are discussed.
Keywords: Biosynthesis; Gibbs energy; Group Contribution Algorithm (GCA); Virtual cell.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Hoehler, T. M., Bains, W., Davila, A., Parenteau, M. N. & Pohorille, A. Life’s requirements, habitability, and biological potential. in (Meadows, V. S., Des Marais, D. J., Arney, G. N. & Schmidt, B. E. Eds.) Planet. Astrobiol. 37–69 (University of Arizona Press, 2020).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

