An across breed, diet and tissue analysis reveals the transcription factor NR1H3 as a key mediator of residual feed intake in beef cattle
- PMID: 38438858
- PMCID: PMC10910725
- DOI: 10.1186/s12864-024-10151-2
An across breed, diet and tissue analysis reveals the transcription factor NR1H3 as a key mediator of residual feed intake in beef cattle
Abstract
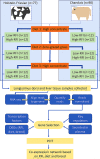
Background: Provision of feed is a major determinant of overall profitability in beef production systems, accounting for up to 75% of the variable costs. Thus, improving cattle feed efficiency, by way of determining the underlying genomic control and subsequently selecting for feed efficient cattle, provides a method through which feed input costs may be reduced. The objective of this study was to undertake gene co-expression network analysis using RNA-Sequence data generated from Longissimus dorsi and liver tissue samples collected from steers of two contrasting breeds (Charolais and Holstein-Friesian) divergent for residual feed intake (RFI), across two consecutive distinct dietary phases (zero-grazed grass and high-concentrate). Categories including differentially expressed genes (DEGs) based on the contrasts of RFI phenotype, breed and dietary source, as well as key transcription factors and proteins secreted in plasma were utilised as nodes of the gene co-expression network.
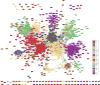
Results: Of the 2,929 DEGs within the network analysis, 1,604 were reported to have statistically significant correlations (≥ 0.80), resulting in a total of 43,876 significant connections between genes. Pathway analysis of clusters of co-expressed genes revealed enrichment of processes related to lipid metabolism (fatty acid biosynthesis, fatty acid β-oxidation, cholesterol biosynthesis), immune function, (complement cascade, coagulation system, acute phase response signalling), and energy production (oxidative phosphorylation, mitochondrial L-carnitine shuttle pathway) based on genes related to RFI, breed and dietary source contrasts.
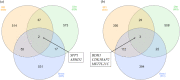
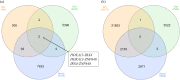
Conclusions: Although similar biological processes were evident across the three factors examined, no one gene node was evident across RFI, breed and diet contrasts in both liver and muscle tissues. However within the liver tissue, the IRX4, NR1H3, HOXA13 and ZNF648 gene nodes, which all encode transcription factors displayed significant connections across the RFI, diet and breed comparisons, indicating a role for these transcription factors towards the RFI phenotype irrespective of diet and breed. Moreover, the NR1H3 gene encodes a protein secreted into plasma from the hepatocytes of the liver, highlighting the potential for this gene to be explored as a robust biomarker for the RFI trait in beef cattle.
Keywords: Beef cattle; Feed efficiency; Gene co-expression network analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The effect of breed and diet type on the global transcriptome of hepatic tissue in beef cattle divergent for feed efficiency.BMC Genomics. 2019 Jun 26;20(1):525. doi: 10.1186/s12864-019-5906-8. BMC Genomics. 2019. PMID: 31242854 Free PMC article.
-
Effect of breed and diet on the M. longissimus thoracis et lumborum transcriptome of steers divergent for residual feed intake.Sci Rep. 2023 Jun 3;13(1):9034. doi: 10.1038/s41598-023-35661-z. Sci Rep. 2023. PMID: 37270611 Free PMC article.
-
Relationship between the rumen microbiome and liver transcriptome in beef cattle divergent for feed efficiency.Anim Microbiome. 2024 Sep 20;6(1):52. doi: 10.1186/s42523-024-00337-0. Anim Microbiome. 2024. PMID: 39304935 Free PMC article.
-
Review: Biological determinants of between-animal variation in feed efficiency of growing beef cattle.Animal. 2018 Dec;12(s2):s321-s335. doi: 10.1017/S1751731118001489. Epub 2018 Aug 24. Animal. 2018. PMID: 30139392 Review.
-
Invited review: Improving feed efficiency of beef cattle - the current state of the art and future challenges.Animal. 2018 Sep;12(9):1815-1826. doi: 10.1017/S1751731118000976. Epub 2018 May 21. Animal. 2018. PMID: 29779496 Review.
Cited by
-
Effects of maternal Escherichia coli lipopolysaccharide exposure on offspring: insights from lncRNA analysis in laying hens.Poult Sci. 2025 Jan;104(1):104599. doi: 10.1016/j.psj.2024.104599. Epub 2024 Nov 30. Poult Sci. 2025. PMID: 39657467 Free PMC article.
References
-
- Bes A, Noziere P, Renand G, Rochette Y, Guarnido-Lopez P, Cantalapiedra-Hijar G, Martin C, et al. Individual methane emissions (and other gas flows) are repeatable and their relationships with feed efficiency are similar across two contrasting diets in growing bulls. Animal. 2022;16:100583. doi: 10.1016/j.animal.2022.100583. - DOI - PubMed
-
- Berry DP, Crowley JJ. Cell biology symposium: genetics of feed efficiency in dairy and beef cattle. J Anim Sci. 2013;91:1594–1613. - PubMed
-
- Fitzsimons C, McGee M, Keogh K, Waters SM, Kenny DA. Molecular physiology of feed efficiency in beef cattle in Biology of Domestic Animals (ed. Hill, R.) CRC Press. 2017;180–231.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

