The RdRp genotyping of SARS-CoV-2 isolated from patients with different clinical spectrum of COVID-19
- PMID: 38439047
- PMCID: PMC10913261
- DOI: 10.1186/s12879-024-09146-x
The RdRp genotyping of SARS-CoV-2 isolated from patients with different clinical spectrum of COVID-19
Abstract
Background: The evolution of SARS-CoV-2 has been observed from the very beginning of the fight against COVID-19, some mutations are indicators of potentially dangerous variants of the virus. However, there is no clear association between the genetic variants of SARS-CoV-2 and the severity of COVID-19. We aimed to analyze the genetic variability of RdRp in correlation with different courses of COVID-19.
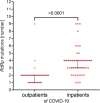
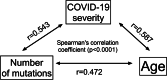
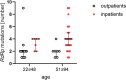
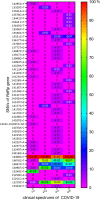
Results: The prospective study included 77 samples of SARS-CoV-2 isolated from outpatients (1st degree of severity) and hospitalized patients (2nd, 3rd and 4th degree of severity). The retrospective analyses included 15,898,266 cases of SARS-CoV-2 genome sequences deposited in the GISAID repository. Single-nucleotide variants were identified based on the four sequenced amplified fragments of SARS-CoV-2. The analysis of the results was performed using appropriate statistical methods, with p < 0.05, considered statistically significant. Additionally, logistic regression analysis was performed to predict the strongest determinants of the observed relationships. The number of mutations was positively correlated with the severity of the COVID-19, and older male patients. We detected four mutations that significantly increased the risk of hospitalization of COVID-19 patients (14676C > T, 14697C > T, 15096 T > C, and 15279C > T), while the 15240C > T mutation was common among strains isolated from outpatients. The selected mutations were searched worldwide in the GISAID database, their presence was correlated with the severity of COVID-19.
Conclusion: Identified mutations have the potential to be used to assess the increased risk of hospitalization in COVID-19 positive patients. Experimental studies and extensive epidemiological data are needed to investigate the association between individual mutations and the severity of COVID-19.
Keywords: RdRp; SARS-CoV-2; Severity of COVID-19.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant.J Transl Med. 2020 Apr 22;18(1):179. doi: 10.1186/s12967-020-02344-6. J Transl Med. 2020. PMID: 32321524 Free PMC article.
-
Evaluation on the use of Nanopore sequencing for direct characterization of coronaviruses from respiratory specimens, and a study on emerging missense mutations in partial RdRP gene of SARS-CoV-2.Virol J. 2020 Nov 23;17(1):183. doi: 10.1186/s12985-020-01454-3. Virol J. 2020. PMID: 33225958 Free PMC article.
-
Different selection dynamics of S and RdRp between SARS-CoV-2 genomes with and without the dominant mutations.Infect Genet Evol. 2021 Jul;91:104796. doi: 10.1016/j.meegid.2021.104796. Epub 2021 Mar 3. Infect Genet Evol. 2021. PMID: 33667722 Free PMC article.
-
Time Series Analysis of SARS-CoV-2 Genomes and Correlations among Highly Prevalent Mutations.Microbiol Spectr. 2022 Oct 26;10(5):e0121922. doi: 10.1128/spectrum.01219-22. Epub 2022 Sep 7. Microbiol Spectr. 2022. PMID: 36069583 Free PMC article.
-
Genomic Characterization of SARS-CoV-2 Isolated from Patients with Distinct Disease Outcomes in Mexico.Microbiol Spectr. 2022 Feb 23;10(1):e0124921. doi: 10.1128/spectrum.01249-21. Epub 2022 Jan 12. Microbiol Spectr. 2022. PMID: 35019701 Free PMC article.
Cited by
-
Biochemical Screening of Phytochemicals and Identification of Scopoletin as a Potential Inhibitor of SARS-CoV-2 Mpro, Revealing Its Biophysical Impact on Structural Stability.Viruses. 2025 Mar 12;17(3):402. doi: 10.3390/v17030402. Viruses. 2025. PMID: 40143329 Free PMC article.
-
Limited Short-Term Evolution of SARS-CoV-2 RNA-Dependent RNA Polymerase under Remdesivir Exposure in Upper Respiratory Compartments.Viruses. 2024 Sep 24;16(10):1511. doi: 10.3390/v16101511. Viruses. 2024. PMID: 39459846 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

