Classification of molecular subtypes for colorectal cancer and development of a prognostic model based on necroptosis-related genes
- PMID: 38439879
- PMCID: PMC10909728
- DOI: 10.1016/j.heliyon.2024.e26781
Classification of molecular subtypes for colorectal cancer and development of a prognostic model based on necroptosis-related genes
Abstract
Background: Necroptosis could regulate immunity in cancers, and stratification of colorectal cancer (CRC) subtypes based on key genes related to necroptosis might be a novel strategy for CRC treatment.
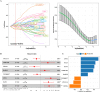
Method: The RNA-sequencing data of CRC and other 31 types of cancers were obtained from The Cancer Genome Atlas (TCGA) database. Consensus clustering was performed based on protein-coding genes (PCGs) related to necroptosis score calculated by single sample gene set enrichment analysis (ssGSEA). Module genes showing a significant positive correlation with the necroptosis score were identified by weighted correlation network analysis (WGCNA) and further used to develop a risk stratification model applying least absolute shrinkage and selection operator (LASSO) and Cox regression analysis. The risks score for each sample in CRC cohorts, immunotherapy cohorts and pan-cancer study cohorts was calculated.
Result: Two subgroups (C1 cluster and C2 cluster) of CRC were identified based on the necroptosis score. Compared with C1 cluster, the survival possibility of C2 cluster was greatly reduced, the levels of necroptosis score, immune cell infiltration, immune score and expression of immune checkpoint molecules were significantly increased and immunotherapy response was less active. Low-risk patients defined by the risk model had a significant survival advantage than high-risk counterparts in both CRC and the other 31 cancer types. Furthermore, the risk model was also more efficient than the Tumor Immune Dysfunction and Exclusion (TIDE) tool in predicting OS and immunotherapy response for the samples in the immunotherapy cohort.
Conclusion: CRC patients were classified by necroptosis score-related PCGs, and a risk model was designed to evaluate the immunotherapy and prognosis of patients with CRC. The current molecular subtype and prognostic model could help stratify patients with different risks and predict their prognosis and immunotherapy sensitivity.
Keywords: Colorectal cancer; Immunotherapy; Molecular subtypes; Necroptosis; Risk stratification tools.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures









References
-
- Siegel R.L., et al. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72(1):7–33. - PubMed
-
- Siegel R.L., et al. Colorectal cancer statistics, 2020. CA A Cancer J. Clin. 2020;70(3):145–164. - PubMed
-
- Arnold M., et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. - PubMed
-
- Harada S., Morlote D. Molecular pathology of colorectal cancer. Adv. Anat. Pathol. 2020;27(1):20–26. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

