WHIRLY protein functions in plants
- PMID: 38440693
- PMCID: PMC10909546
- DOI: 10.1002/fes3.379
WHIRLY protein functions in plants
Abstract
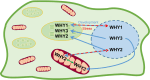
Environmental stresses pose a significant threat to food security. Understanding the function of proteins that regulate plant responses to biotic and abiotic stresses is therefore pivotal in developing strategies for crop improvement. The WHIRLY (WHY) family of DNA-binding proteins are important in this regard because they fulfil a portfolio of important functions in organelles and nuclei. The WHY1 and WHY2 proteins function as transcription factors in the nucleus regulating phytohormone synthesis and associated growth and stress responses, as well as fulfilling crucial roles in DNA and RNA metabolism in plastids and mitochondria. WHY1, WHY2 (and WHY3 proteins in Arabidopsis) maintain organelle genome stability and serve as auxiliary factors for homologous recombination and double-strand break repair. Our understanding of WHY protein functions has greatly increased in recent years, as has our knowledge of the flexibility of their localization and overlap of functions but there is no review of the topic in the literature. Our aim in this review was therefore to provide a comprehensive overview of the topic, discussing WHY protein functions in nuclei and organelles and highlighting roles in plant development and stress responses. In particular, we consider areas of uncertainty such as the flexible localization of WHY proteins in terms of retrograde signalling connecting mitochondria, plastids, and the nucleus. Moreover, we identify WHY proteins as important targets in plant breeding programmes designed to increase stress tolerance and the sustainability of crop yield in a changing climate.
Keywords: DNA damage; chloroplasts; homologous recombination; mitochondria; nucleus; pathogenesis‐related genes.
© 2022 The Authors. Food and Energy Security published by John Wiley & Sons Ltd.
Conflict of interest statement
There are no conflicting interests.
Figures


References
-
- Bawono, P. , & Heringa, J. (2014). PRALINE: A versatile multiple sequence alignment toolkit. Methods in Molecular Biology, 1079, 245–262. - PubMed
-
- Cappadocia, L. , Parent, J. S. , Sygusch, J. , & Brisson, N. (2013). A family portrait: structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum. Acta crystallographica . Section F, Structural Biology and Crystallization Communications, 69, 1207–1211. 10.1107/S1744309113028698 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
