Post-translational modifications of proteins in cardiovascular diseases examined by proteomic approaches
- PMID: 38440918
- PMCID: PMC11705224
- DOI: 10.1111/febs.17108
Post-translational modifications of proteins in cardiovascular diseases examined by proteomic approaches
Abstract
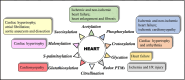
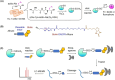
Over 400 different types of post-translational modifications (PTMs) have been reported and over 200 various types of PTMs have been discovered using mass spectrometry (MS)-based proteomics. MS-based proteomics has proven to be a powerful method capable of global PTM mapping with the identification of modified proteins/peptides, the localization of PTM sites and PTM quantitation. PTMs play regulatory roles in protein functions, activities and interactions in various heart related diseases, such as ischemia/reperfusion injury, cardiomyopathy and heart failure. The recognition of PTMs that are specific to cardiovascular pathology and the clarification of the mechanisms underlying these PTMs at molecular levels are crucial for discovery of novel biomarkers and application in a clinical setting. With sensitive MS instrumentation and novel biostatistical methods for precise processing of the data, low-abundance PTMs can be successfully detected and the beneficial or unfavorable effects of specific PTMs on cardiac function can be determined. Moreover, computational proteomic strategies that can predict PTM sites based on MS data have gained an increasing interest and can contribute to characterization of PTM profiles in cardiovascular disorders. More recently, machine learning- and deep learning-based methods have been employed to predict the locations of PTMs and explore PTM crosstalk. In this review article, the types of PTMs are briefly overviewed, approaches for PTM identification/quantitation in MS-based proteomics are discussed and recently published proteomic studies on PTMs associated with cardiovascular diseases are included.
Keywords: MS‐based proteomics; cardiovascular disease; post‐translational modifications; proteins.
© 2024 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The author declares that he has no conflicts of interest.
Figures


Similar articles
-
The Methods Employed in Mass Spectrometric Analysis of Posttranslational Modifications (PTMs) and Protein-Protein Interactions (PPIs).Adv Exp Med Biol. 2019;1140:169-198. doi: 10.1007/978-3-030-15950-4_10. Adv Exp Med Biol. 2019. PMID: 31347048 Free PMC article. Review.
-
Substrate and Functional Diversity of Protein Lysine Post-translational Modifications.Genomics Proteomics Bioinformatics. 2024 May 9;22(1):qzae019. doi: 10.1093/gpbjnl/qzae019. Genomics Proteomics Bioinformatics. 2024. PMID: 38862432 Free PMC article. Review.
-
dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications.Nucleic Acids Res. 2019 Jan 8;47(D1):D298-D308. doi: 10.1093/nar/gky1074. Nucleic Acids Res. 2019. PMID: 30418626 Free PMC article.
-
Post-translational quantitation by SRM/MRM: applications in cardiology.Expert Rev Proteomics. 2018 Jun;15(6):477-502. doi: 10.1080/14789450.2018.1484283. Expert Rev Proteomics. 2018. PMID: 29865883 Review.
-
Identification, Quantification, and Site Localization of Protein Posttranslational Modifications via Mass Spectrometry-Based Proteomics.Adv Exp Med Biol. 2016;919:345-382. doi: 10.1007/978-3-319-41448-5_17. Adv Exp Med Biol. 2016. PMID: 27975226 Review.
Cited by
-
Decoding glycosylation in cardiovascular diseases: mechanisms, biomarkers, and therapeutic opportunities.Front Pharmacol. 2025 May 19;16:1570158. doi: 10.3389/fphar.2025.1570158. eCollection 2025. Front Pharmacol. 2025. PMID: 40458788 Free PMC article. Review.
-
A Systematic Study of Lysine Succinylation in the Pathogenic Bacterium Vibrio harveyi in Aquatic Animals.Molecules. 2025 May 31;30(11):2418. doi: 10.3390/molecules30112418. Molecules. 2025. PMID: 40509307 Free PMC article.
-
Acetylation of Steroidogenic Acute Regulatory Protein Sensitizes 17β-Estradiol Regulation in Hormone-Sensitive Breast Cancer Cells.Int J Mol Sci. 2024 Aug 10;25(16):8732. doi: 10.3390/ijms25168732. Int J Mol Sci. 2024. PMID: 39201419 Free PMC article.
-
The Role of Proteomics in Identification of Key Proteins of Bacterial Cells with Focus on Probiotic Bacteria.Int J Mol Sci. 2024 Aug 6;25(16):8564. doi: 10.3390/ijms25168564. Int J Mol Sci. 2024. PMID: 39201251 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

