Complete nucleotide sequence and comparative genomic analysis of microcin B17 plasmid pMccB17
- PMID: 38440924
- PMCID: PMC10912980
- DOI: 10.1002/mbo3.1402
Complete nucleotide sequence and comparative genomic analysis of microcin B17 plasmid pMccB17
Abstract
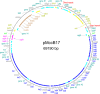
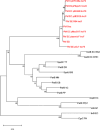
We present a comprehensive sequence and bioinformatic analysis of the prototypical microcin plasmid, pMccb17, which includes a definitive sequence for the microcin operon, mcb. Microcin B17 (MccB17) is a ribosomally synthesized and posttranslationally modified peptide produced by Escherichia coli. It inhibits bacterial DNA gyrase similarly to quinolone antibiotics. The mcb operon, which consists of seven genes encoding biosynthetic and immunity/export functions, was originally located on the low copy number IncFII plasmid pMccB17 in the Escherichia coli strain LP17. It was later transferred to E. coli K-12 through conjugation. In this study, the plasmid was extracted from the E. coli K-12 strain RYC1000 [pMccB17] and sequenced twice using an Illumina short-read method. The first sequencing was conducted with the host bacterial chromosome, and the plasmid DNA was then purified and sequenced separately. After assembly into a single contig, polymerase chain reaction primers were designed to close the single remaining gap via Sanger sequencing. The resulting complete circular DNA sequence is 69,190 bp long and includes 81 predicted genes. These genes were initially identified by Prokka and subsequently manually reannotated using BLAST. The plasmid was assigned to the F2:A-:B- replicon type with a MOBF12 group conjugation system. A comparison with other IncFII plasmids revealed a large proportion of shared genes, particularly in the conjugative plasmid backbone. However, unlike many contemporary IncFII plasmids, pMccB17 lacks transposable elements and antibiotic resistance genes. In addition to the mcb operon, this plasmid carries 25 genes of unknown function.
Keywords: Enterobacteriaceae; genome; microcin; plasmid.
© 2024 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Cloning and mapping of the genetic determinants for microcin B17 production and immunity.J Bacteriol. 1985 Jul;163(1):275-81. doi: 10.1128/jb.163.1.275-281.1985. J Bacteriol. 1985. PMID: 2989247 Free PMC article.
-
Microcin B17, a novel tool for preparation of maxicells: identification of polypeptides encoded by the IncFII minireplicon pMccB17.J Bacteriol. 1988 May;170(5):2414-7. doi: 10.1128/jb.170.5.2414-2417.1988. J Bacteriol. 1988. PMID: 3283111 Free PMC article.
-
Evidence that colicin X is microcin B17.J Bacteriol. 1987 Jun;169(6):2899-901. doi: 10.1128/jb.169.6.2899-2901.1987. J Bacteriol. 1987. PMID: 3034869 Free PMC article.
-
The structural gene for microcin H47 encodes a peptide precursor with antibiotic activity.Antimicrob Agents Chemother. 1999 Sep;43(9):2176-82. doi: 10.1128/AAC.43.9.2176. Antimicrob Agents Chemother. 1999. PMID: 10471561 Free PMC article. Review.
-
The Microbial Toxin Microcin B17: Prospects for the Development of New Antibacterial Agents.J Mol Biol. 2019 Aug 23;431(18):3400-3426. doi: 10.1016/j.jmb.2019.05.050. Epub 2019 Jun 8. J Mol Biol. 2019. PMID: 31181289 Free PMC article. Review.
References
-
- Arnison, P. G. , Bibb, M. J. , Bierbaum, G. , Bowers, A. A. , Bugni, T. S. , Bulaj, G. , Camarero, J. A. , Campopiano, D. J. , Challis, G. L. , Clardy, J. , Cotter, P. D. , Craik, D. J. , Dawson, M. , Dittmann, E. , Donadio, S. , Dorrestein, P. C. , Entian, K. D. , Fischbach, M. A. , Garavelli, J. S. , … van der Donk, W. A. (2013). Ribosomally synthesized and post‐translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Natural Products Reports, 30(1), 108–160. 10.1039/C2NP20085F - DOI - PMC - PubMed
-
- Bankevich, A. , Nurk, S. , Antipov, D. , Gurevich, A. A. , Dvorkin, M. , Kulikov, A. S. , Lesin, V. M. , Nikolenko, S. I. , Pham, S. , Prjibelski, A. D. , Pyshkin, A. V. , Sirotkin, A. V. , Vyahhi, N. , Tesler, G. , Alekseyev, M. A. , & Pevzner, P. A. (2012). SPAdes: a new genome assembly algorithm and its applications to single‐cell sequencing. Journal of Computational Biology, 19(5), 455–477. 10.1089/cmb.2012.0021 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

