FeLIX is a restriction factor for mammalian retrovirus infection
- PMID: 38440982
- PMCID: PMC11019853
- DOI: 10.1128/jvi.01771-23
FeLIX is a restriction factor for mammalian retrovirus infection
Abstract
Endogenous retroviruses (ERVs) are remnants of ancestral viral infections. Feline leukemia virus (FeLV) is an exogenous and endogenous retrovirus in domestic cats. It is classified into several subgroups (A, B, C, D, E, and T) based on viral receptor interference properties or receptor usage. ERV-derived molecules benefit animals, conferring resistance to infectious diseases. However, the soluble protein encoded by the defective envelope (env) gene of endogenous FeLV (enFeLV) functions as a co-factor in FeLV subgroup T infections. Therefore, whether the gene emerged to facilitate viral infection is unclear. Based on the properties of ERV-derived molecules, we hypothesized that the defective env genes possess antiviral activity that would be advantageous to the host because FeLV subgroup B (FeLV-B), a recombinant virus derived from enFeLV env, is restricted to viral transmission among domestic cats. When soluble truncated Env proteins from enFeLV were tested for their inhibitory effects against enFeLV and FeLV-B, they inhibited viral infection. Notably, this antiviral machinery was extended to infection with the Gibbon ape leukemia virus, Koala retrovirus A, and Hervey pteropid gammaretrovirus. Although these viruses used feline phosphate transporter 1 (fePit1) and phosphate transporter 2 as receptors, the inhibitory mechanism involved competitive receptor binding in a fePit1-dependent manner. The shift in receptor usage might have occurred to avoid the inhibitory effect. Overall, these findings highlight the possible emergence of soluble truncated Env proteins from enFeLV as a restriction factor against retroviral infection and will help in developing host immunity and antiviral defense by controlling retroviral spread.IMPORTANCERetroviruses are unique in using reverse transcriptase to convert RNA genomes into DNA, infecting germ cells, and transmitting to offspring. Numerous ancient retroviral sequences are known as endogenous retroviruses (ERVs). The soluble Env protein derived from ERVs functions as a co-factor that assists in FeLV-T infection. However, herein, we show that the soluble Env protein exhibits antiviral activity and provides resistance to mammalian retrovirus infection through competitive receptor binding. In particular, this finding may explain why FeLV-B transmission is not observed among domestic cats. ERV-derived molecules can benefit animals in an evolutionary arms race, highlighting the double-edged-sword nature of ERVs.
Keywords: endogenous retrovirus; feline leukemia virus; restriction factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



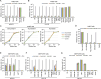
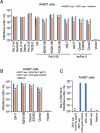
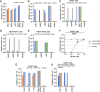
References
-
- Coffin JM, Hughes SH, Varmus HE. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY). - PubMed
-
- Miyake A, Ngo MH, Wulandari S, Shimojima M, Nakagawa S, Kawasaki J, Nishigaki K. 2022. Convergent evolution of antiviral machinery derived from endogenous retrovirus truncated envelope genes in multiple species. Proc Natl Acad Sci U S A 119:e2114441119. doi:10.1073/pnas.2114441119 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

