An inquiline mosquito modulates microbial diversity and function in an aquatic microecosystem
- PMID: 38441172
- PMCID: PMC10989397
- DOI: 10.1111/mec.17314
An inquiline mosquito modulates microbial diversity and function in an aquatic microecosystem
Abstract
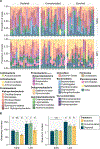
Understanding microbial roles in ecosystem function requires integrating microscopic processes into food webs. The carnivorous pitcher plant, Sarracenia purpurea, offers a tractable study system where diverse food webs of macroinvertebrates and microbes facilitate digestion of captured insect prey, releasing nutrients supporting the food web and host plant. However, how interactions between these macroinvertebrate and microbial communities contribute to ecosystem functions remains unclear. We examined the role of the pitcher plant mosquito, Wyeomyia smithii, in top-down control of the composition and function of pitcher plant microbial communities. Mosquito larval abundance was enriched or depleted across a natural population of S. purpurea pitchers over a 74-day field experiment. Bacterial community composition and microbial community function were characterized by 16S rRNA amplicon sequencing and profiling of carbon substrate use, bulk metabolic rate, hydrolytic enzyme activity, and macronutrient pools. Bacterial communities changed from pitcher opening to maturation, but larvae exerted minor effects on high-level taxonomic composition. Higher larval abundance was associated with lower diversity communities with distinct functions and elevated nitrogen availability. Treatment-independent clustering also supported roles for larvae in curating pitcher microbial communities through shifts in community diversity and function. These results demonstrate top-down control of microbial functions in an aquatic microecosystem.
Keywords: carnivorous plant; microbiota; nitrogen; nutrient degradation/cycling; symbiosis; top‐down control.
© 2024 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
Conflicts of interest
All authors declare no conflicts of interest.
Figures




References
-
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto J-M, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, … Bork P (2011). Enterotypes of the human gut microbiome. Nature, 473(7346), 174–180. 10.1038/nature09944 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

