Uncertainty-aware single-cell annotation with a hierarchical reject option
- PMID: 38441258
- PMCID: PMC10957513
- DOI: 10.1093/bioinformatics/btae128
Uncertainty-aware single-cell annotation with a hierarchical reject option
Abstract
Motivation: Automatic cell type annotation methods assign cell type labels to new datasets by extracting relationships from a reference RNA-seq dataset. However, due to the limited resolution of gene expression features, there is always uncertainty present in the label assignment. To enhance the reliability and robustness of annotation, most machine learning methods address this uncertainty by providing a full reject option, i.e. when the predicted confidence score of a cell type label falls below a user-defined threshold, no label is assigned and no prediction is made. As a better alternative, some methods deploy hierarchical models and consider a so-called partial rejection by returning internal nodes of the hierarchy as label assignment. However, because a detailed experimental analysis of various rejection approaches is missing in the literature, there is currently no consensus on best practices.
Results: We evaluate three annotation approaches (i) full rejection, (ii) partial rejection, and (iii) no rejection for both flat and hierarchical probabilistic classifiers. Our findings indicate that hierarchical classifiers are superior when rejection is applied, with partial rejection being the preferred rejection approach, as it preserves a significant amount of label information. For optimal rejection implementation, the rejection threshold should be determined through careful examination of a method's rejection behavior. Without rejection, flat and hierarchical annotation perform equally well, as long as the cell type hierarchy accurately captures transcriptomic relationships.
Availability and implementation: Code is freely available at https://github.com/Latheuni/Hierarchical_reject and https://doi.org/10.5281/zenodo.10697468.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
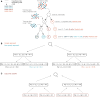
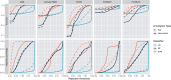

References
-
- Beygelzimer A, Langford J, Lifshits Y. et al. Conditional probability tree estimation analysis and algorithms. In: Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence, UAI ’09, Arlington, Virginia, United States. AUAI Press, 2009, 51–8.
-
- Bi W, Kwok JT.. Bayes-optimal hierarchical multilabel classification. IEEE Trans Knowl Data Eng 2015;27:2907–18.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

