Whole-genome sequencing-based antimicrobial resistance and shedding dynamics of Escherichia coli isolated from calves before and after antimicrobial group treatments
- PMID: 38441467
- PMCID: PMC10986552
- DOI: 10.1128/spectrum.03214-23
Whole-genome sequencing-based antimicrobial resistance and shedding dynamics of Escherichia coli isolated from calves before and after antimicrobial group treatments
Abstract
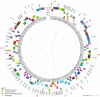
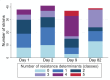
The fattening of calves is often associated with high antimicrobial use and the selection of antimicrobial resistance (AMR). The objective of this observational longitudinal study was to describe the AMR and strain dynamics, using whole-genome sequencing (WGS), of fecal Escherichia coli in a cohort of 22 calves. All calves received antimicrobial group treatments on Day (D) 1 (oxytetracycline, intramuscularly) and on D4 through D12 (doxycycline, in-feed). Additionally, eight calves received individual parenteral treatments between D7 and D59, including florfenicol, amoxicillin, marbofloxacin, and gamithromycin. Rectal swabs were collected from all calves on D1 (prior to treatment), D2, D9, and D82. The swabs were spread onto Enterobacterales-selective agar, and three E. coli colonies per plate were subjected to WGS. Out of 264 isolates across all calves and sampling times, 80 unique strains were identified, a majority of which harbored genes conferring resistance to tetracyclines, streptomycin, and sulfonamides. The diversity of strains decreased during the in-feed antimicrobial group treatment of the calves. On D82, 90% of isolates were strains that were not isolated at previous sampling times, and the median number per strain of AMR determinants to tetracyclines, florfenicol, β-lactams, quinolones, or macrolides decreased compared to D9. Additionally, clonal dissemination of some strains represented the main transmission route of AMR determinants. In this study, WGS revealed important variations in strain diversity and genotypic AMR of fecal E. coli over time in calves subjected to group antimicrobial treatments.
Importance: The continued emergence and spread of antimicrobial resistance (AMR) determinants are serious global concerns. The dynamics of AMR spread and persistence in bacterial and animal host populations are complex and not solely driven by antimicrobial selection pressure. In calf fattening, both antimicrobial use and carriage prevalence of antimicrobial-resistant bacteria are generally recognized as high. This study provides new insights into the short-term, within-farm dynamics and transmission of AMR determinants in Escherichia coli from the dominant fecal flora of calves subjected to antimicrobial group treatments during the rearing period. The diversity of E. coli strains decreased over time, although, in contrast to previous observations in extended-spectrum β-lactamase-producing Enterobacterales, the predominance of a few clones was not observed. The spread of AMR determinants occurred through the dissemination of clonal strains among calves. The median number per strain of AMR determinants conferring resistance to selected antimicrobials decreased toward the end of the rearing period.
Keywords: Escherichia coli; antimicrobial resistance; cattle; fecal carriage; molecular epidemiology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Effects of antimicrobial exposure on the antimicrobial resistance of Escherichia coli in the digestive flora of dairy calves.Prev Vet Med. 2020 Dec;185:105177. doi: 10.1016/j.prevetmed.2020.105177. Epub 2020 Oct 10. Prev Vet Med. 2020. PMID: 33181469
-
Fecal Carriage and Whole-Genome Sequencing-Assisted Characterization of CMY-2 Beta-Lactamase-Producing Escherichia coli in Calves at Czech Dairy Cow Farm.Foodborne Pathog Dis. 2019 Jan;16(1):42-53. doi: 10.1089/fpd.2018.2531. Foodborne Pathog Dis. 2019. PMID: 30673354
-
Antimicrobial resistance profiles of Escherichia coli and prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in calves from organic and conventional dairy farms in Switzerland.Microbiologyopen. 2022 Apr;11(2):e1269. doi: 10.1002/mbo3.1269. Microbiologyopen. 2022. PMID: 35478290 Free PMC article.
-
Antimicrobial Resistance in Fecal Escherichia coli and Salmonella enterica from Dairy Calves: A Systematic Review.Foodborne Pathog Dis. 2019 Jan;16(1):23-34. doi: 10.1089/fpd.2018.2529. Epub 2018 Nov 27. Foodborne Pathog Dis. 2019. PMID: 30481058
-
A scoping review of antimicrobial resistance in the Australian dairy cattle industry.Prev Vet Med. 2024 May;226:106161. doi: 10.1016/j.prevetmed.2024.106161. Epub 2024 Feb 24. Prev Vet Med. 2024. PMID: 38460345
Cited by
-
Dairy Cattle and the Iconic Autochthonous Cattle in Northern Portugal Are Reservoirs of Multidrug-Resistant Escherichia coli.Antibiotics (Basel). 2024 Dec 11;13(12):1208. doi: 10.3390/antibiotics13121208. Antibiotics (Basel). 2024. PMID: 39766598 Free PMC article.
References
-
- Lava M, Pardon B, Schüpbach-Regula G, Keckeis K, Deprez P, Steiner A, Meylan M. 2016. Effect of calf purchase and other herd-level risk factors on mortality, unwanted early slaughter, and use of antimicrobial group treatments in Swiss veal calf operations. Prev Vet Med 126:81–88. doi:10.1016/j.prevetmed.2016.01.020 - DOI - PubMed
-
- Catry B, Dewulf J, Maes D, Pardon B, Callens B, Vanrobaeys M, Opsomer G, de Kruif A, Haesebrouck F. 2016. Effect of antimicrobial consumption and production type on antibacterial resistance in the bovine respiratory and digestive tract. PLoS One 11:e0146488. doi:10.1371/journal.pone.0146488 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

