Comparative analysis of IR-Biotyper, MLST, cgMLST, and WGS for clustering of vancomycin-resistant Enterococcus faecium in a neonatal intensive care unit
- PMID: 38441473
- PMCID: PMC10986520
- DOI: 10.1128/spectrum.04119-23
Comparative analysis of IR-Biotyper, MLST, cgMLST, and WGS for clustering of vancomycin-resistant Enterococcus faecium in a neonatal intensive care unit
Abstract
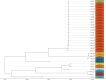
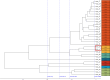
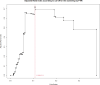
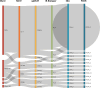
Healthcare-associated infections caused by vancomycin-resistant Enterococcus faecium (VREFM) pose a significant threat to healthcare. Confirming the relatedness of the bacterial isolates from different patients is challenging. We aimed to assess the efficacy of IR-Biotyper, multilocus sequencing typing (MLST), and core-genome MLST (cgMLST) in comparison with whole-genome sequencing (WGS) for outbreak confirmation in the neonatal intensive care unit (NICU). Twenty VREFM isolates from four neonates and ten control isolates from unrelated patients were analyzed. Genomic DNA extraction, MLST, cgMLST, and WGS were performed. An IR-Biotyper was used with colonies obtained after 24 h of incubation on tryptic soy agar supplemented with 5% sheep blood. The optimal clustering cutoff for the IR-Biotyper was determined by comparing the results with WGS. Clustering concordance was assessed using the adjusted Rand and Wallace indices. MLST and cgMLST identified sequence types (ST) and complex types (CT), revealing suspected outbreak isolates with a predominance of ST17 and CT6553, were confirmed by WGS. For the IR-Biotyper, the proposed optimal clustering cut-off range was 0.106-0.111. Despite lower within-run precision, of the IR-Biotyper, the clustering concordance with WGS was favorable, meeting the criteria for real-time screening. This study confirmed a nosocomial outbreak of VREFM in the NICU using an IR-Biotyper, showing promising results compared to MLST. Although within-run precision requires improvement, the IR-Biotyper demonstrated high discriminatory power and clustering concordance with WGS. These findings suggest its potential as a real-time screening tool for the detection of VREFM-related nosocomial outbreaks.
Importance: In this study, we evaluated the performance of the IR-Biotyper in detecting nosocomial outbreaks caused by vancomycin-resistant Enterococcus faecium, comparing it with MLST, cgMLST, and WGS. We proposed a cutoff that showed the highest concordance compared to WGS and assessed the within-run precision of the IR-Biotyper by evaluating the consistency in genetically identical strain when repeated in the same run.
Keywords: IR-Biotyper; VREFM; nosocomial outbreak.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Development of a strain-specific PCR as a diagnostic tool for surveillance, detection, and monitoring of vancomycin-resistant Enterococcus faecium during outbreak.Antimicrob Resist Infect Control. 2025 Mar 24;14(1):23. doi: 10.1186/s13756-025-01538-1. Antimicrob Resist Infect Control. 2025. PMID: 40128808 Free PMC article.
-
New multilocus sequence typing scheme for Enterococcus faecium reveals sequential outbreaks of vancomycin-resistant E. faecium ST1162 and ST610 in a Japanese tertiary medical center.Microbiol Spectr. 2025 Jan 7;13(1):e0213124. doi: 10.1128/spectrum.02131-24. Epub 2024 Dec 10. Microbiol Spectr. 2025. PMID: 39656011 Free PMC article.
-
Early identification of the nosocomial spread of vancomycin-resistant Enterococcus faecium by Fourier-transform infrared spectroscopy and performance comparison with PFGE and WGS.Emerg Microbes Infect. 2024 Dec;13(1):2392659. doi: 10.1080/22221751.2024.2392659. Epub 2024 Aug 23. Emerg Microbes Infect. 2024. PMID: 39137261 Free PMC article.
-
The use of core genome multilocus sequence typing to determine the duration of vancomycin-resistant Enterococcus faecium outbreaks.APMIS. 2022 Jun;130(6):323-329. doi: 10.1111/apm.13216. Epub 2022 Mar 31. APMIS. 2022. PMID: 35253272 Review.
-
A molecular study regarding the spread of vanA vancomycin-resistant Enterococcus faecium in a tertiary hospital in China.J Glob Antimicrob Resist. 2022 Dec;31:270-278. doi: 10.1016/j.jgar.2022.10.010. Epub 2022 Oct 20. J Glob Antimicrob Resist. 2022. PMID: 36273808 Review.
Cited by
-
Fourier-transform infrared spectroscopy for rapid Streptococcus pneumoniae serotyping in a tertiary care general hospital.Front Microbiol. 2025 Apr 23;16:1565888. doi: 10.3389/fmicb.2025.1565888. eCollection 2025. Front Microbiol. 2025. PMID: 40336825 Free PMC article.
-
Development of a robust FT-IR typing system for Salmonella enterica, enhancing performance through hierarchical classification.Microbiol Spectr. 2025 Jul;13(7):e0015925. doi: 10.1128/spectrum.00159-25. Epub 2025 May 27. Microbiol Spectr. 2025. PMID: 40422737 Free PMC article.
References
-
- Teng ASJ, Habermehl PE, van Houdt R, de Jong MD, van Mansfeld R, Matamoros SPF, Spijkerman IJB, van Meer MPA, Visser CE. 2022. Comparison of fast fourier transform infrared spectroscopy biotyping with whole genome sequencing-based genotyping in common nosocomial pathogens. Anal Bioanal Chem 414:7179–7189. doi:10.1007/s00216-022-04270-6 - DOI - PMC - PubMed
-
- Higgs C, Sherry NL, Seemann T, Horan K, Walpola H, Kinsella P, Bond K, Williamson DA, Marshall C, Kwong JC, Grayson ML, Stinear TP, Gorrie CL, Howden BP. 2022. Optimising genomic approaches for identifying vancomycin-resistant Enterococcus faecium transmission in healthcare settings. Nat Commun 13:509. doi:10.1038/s41467-022-28156-4 - DOI - PMC - PubMed
-
- Rakovitsky N, Frenk S, Kon H, Schwartz D, Temkin E, Solter E, Paikin S, Cohen R, Schwaber MJ, Carmeli Y, Lellouche J. 2020. Fourier transform infrared spectroscopy is a new option for outbreak investigation: a retrospective analysis of an extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. J Clin Microbiol 58:e00098-20. doi:10.1128/JCM.00098-20 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

