Trinucleotide mRNA Cap Analogue N 6-Benzylated at the Site of Posttranscriptional m6Am Mark Facilitates mRNA Purification and Confers Superior Translational Properties In Vitro and In Vivo
- PMID: 38442005
- PMCID: PMC10979456
- DOI: 10.1021/jacs.3c12629
Trinucleotide mRNA Cap Analogue N 6-Benzylated at the Site of Posttranscriptional m6Am Mark Facilitates mRNA Purification and Confers Superior Translational Properties In Vitro and In Vivo
Abstract
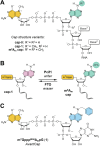
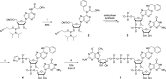
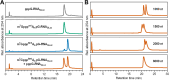
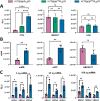
Eukaryotic mRNAs undergo cotranscriptional 5'-end modification with a 7-methylguanosine cap. In higher eukaryotes, the cap carries additional methylations, such as m6Am─a common epitranscriptomic mark unique to the mRNA 5'-end. This modification is regulated by the Pcif1 methyltransferase and the FTO demethylase, but its biological function is still unknown. Here, we designed and synthesized a trinucleotide FTO-resistant N6-benzyl analogue of the m6Am-cap-m7GpppBn6AmpG (termed AvantCap) and incorporated it into mRNA using T7 polymerase. mRNAs carrying Bn6Am showed several advantages over typical capped transcripts. The Bn6Am moiety was shown to act as a reversed-phase high-performance liquid chromatography (RP-HPLC) purification handle, allowing the separation of capped and uncapped RNA species, and to produce transcripts with lower dsRNA content than reference caps. In some cultured cells, Bn6Am mRNAs provided higher protein yields than mRNAs carrying Am or m6Am, although the effect was cell-line-dependent. m7GpppBn6AmpG-capped mRNAs encoding reporter proteins administered intravenously to mice provided up to 6-fold higher protein outputs than reference mRNAs, while mRNAs encoding tumor antigens showed superior activity in therapeutic settings as anticancer vaccines. The biochemical characterization suggests several phenomena potentially underlying the biological properties of AvantCap: (i) reduced propensity for unspecific interactions, (ii) involvement in alternative translation initiation, and (iii) subtle differences in mRNA impurity profiles or a combination of these effects. AvantCapped-mRNAs bearing the Bn6Am may pave the way for more potent mRNA-based vaccines and therapeutics and serve as molecular tools to unravel the role of m6Am in mRNA.
Conflict of interest statement
The authors declare the following competing financial interest(s): J.J., J.K., P.J.S., and M.W. are inventors of a patent related to AvantCap. Some of the authors are shareholders of Explorna Therapuetics.
Figures



Similar articles
-
Enzymatic characterization of mRNA cap adenosine-N6 methyltransferase PCIF1 activity on uncapped RNAs.J Biol Chem. 2022 Apr;298(4):101751. doi: 10.1016/j.jbc.2022.101751. Epub 2022 Feb 19. J Biol Chem. 2022. PMID: 35189146 Free PMC article.
-
Synthesis of biotin labelled cap analogue--incorporable into mRNA transcripts and promoting cap-dependent translation.Org Biomol Chem. 2012 Nov 21;10(43):8570-4. doi: 10.1039/c2ob26060c. Org Biomol Chem. 2012. PMID: 22832840
-
The identity and methylation status of the first transcribed nucleotide in eukaryotic mRNA 5' cap modulates protein expression in living cells.Nucleic Acids Res. 2020 Feb 28;48(4):1607-1626. doi: 10.1093/nar/gkaa032. Nucleic Acids Res. 2020. PMID: 31984425 Free PMC article.
-
Cap-Independent Translation: What's in a Name?Trends Biochem Sci. 2018 Nov;43(11):882-895. doi: 10.1016/j.tibs.2018.04.011. Epub 2018 May 19. Trends Biochem Sci. 2018. PMID: 29789219 Review.
-
RNA methyltransferases involved in 5' cap biosynthesis.RNA Biol. 2014;11(12):1597-607. doi: 10.1080/15476286.2015.1004955. RNA Biol. 2014. PMID: 25626080 Free PMC article. Review.
Cited by
-
5' terminal nucleotide determines the immunogenicity of IVT RNAs.Nucleic Acids Res. 2025 Jan 24;53(3):gkae1252. doi: 10.1093/nar/gkae1252. Nucleic Acids Res. 2025. PMID: 39704128 Free PMC article.
-
Recent Advancements in mRNA Vaccines: From Target Selection to Delivery Systems.Vaccines (Basel). 2024 Aug 1;12(8):873. doi: 10.3390/vaccines12080873. Vaccines (Basel). 2024. PMID: 39203999 Free PMC article. Review.
-
Discovery and development of a safe and efficient COVID-19 mRNA vaccine, STP2104, using a novel capping library screening method.Front Immunol. 2025 Jun 9;16:1571713. doi: 10.3389/fimmu.2025.1571713. eCollection 2025. Front Immunol. 2025. PMID: 40552291 Free PMC article.
-
mRNA medicine: Recent progresses in chemical modification, design, and engineering.Nano Res. 2024 Oct;17(10):9015-9030. doi: 10.1007/s12274-024-6978-6. Epub 2024 Sep 3. Nano Res. 2024. PMID: 40756674
-
Application of Mammalian Nudix Enzymes to Capped RNA Analysis.Pharmaceuticals (Basel). 2024 Sep 11;17(9):1195. doi: 10.3390/ph17091195. Pharmaceuticals (Basel). 2024. PMID: 39338357 Free PMC article. Review.
References
-
- Furuichi Y.; Muthukrishnan S.; Shatkin A. J. 5′-Terminal m-7G(5′)ppp(5′)G-m-p in vivo: identification in reovirus genome RNA. Proc. Natl. Acad. Sci. U.S.A. 1975, 72 (2), 742–745. 10.1073/pnas.72.2.742. - DOI - PMC - PubMed
- Bélanger F.; Stepinski J.; Darzynkiewicz E.; Pelletier J. Characterization of hMTr1, a Human Cap1 2′-O-Ribose Methyltransferase*. J. Biol. Chem. 2010, 285 (43), 33037–33044. 10.1074/jbc.M110.155283. - DOI - PMC - PubMed
- Langberg S. R.; Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2′-)-methyltransferases from HeLa cells. J. Biol. Chem. 1981, 256 (19), 10054–10060. 10.1016/S0021-9258(19)68740-5. - DOI - PubMed
-
- Drazkowska K.; Tomecki R.; Warminski M.; Baran N.; Cysewski D.; Depaix A.; Kasprzyk R.; Kowalska J.; Jemielity J.; Sikorski P. J. 2′-O-Methylation of the second transcribed nucleotide within the mRNA 5′ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 2022, 50 (16), 9051–9071. 10.1093/nar/gkac722. - DOI - PMC - PubMed
-
- Werner M.; Purta E.; Kaminska K. H.; Cymerman I. A.; Campbell D. A.; Mittra B.; Zamudio J. R.; Sturm N. R.; Jaworski J.; Bujnicki J. M. 2′-O-ribose methylation of cap2 in human: function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011, 39 (11), 4756–4768. 10.1093/nar/gkr038. - DOI - PMC - PubMed
- Despic V.; Jaffrey S. R. mRNA ageing shapes the Cap2 methylome in mammalian mRNA. Nature 2023, 614 (7947), 358–366. 10.1038/s41586-022-05668-z. - DOI - PMC - PubMed
- Smietanski M.; Werner M.; Purta E.; Kaminska K. H.; Stepinski J.; Darzynkiewicz E.; Nowotny M.; Bujnicki J. M. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat. Commun. 2014, 5, 300410.1038/ncomms4004. - DOI - PMC - PubMed
-
- Sahin U.; Muik A.; Derhovanessian E.; Vogler I.; Kranz L. M.; Vormehr M.; Baum A.; Pascal K.; Quandt J.; Maurus D.; et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 2020, 586 (7830), 594–599. 10.1038/s41586-020-2814-7. - DOI - PubMed
- Corbett K. S.; Edwards D. K.; Leist S. R.; Abiona O. M.; Boyoglu-Barnum S.; Gillespie R. A.; Himansu S.; Schäfer A.; Ziwawo C. T.; DiPiazza A. T.; et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature 2020, 586 (7830), 567–571. 10.1038/s41586-020-2622-0. - DOI - PMC - PubMed
-
- Wei C.-M.; Gershowitz A.; Moss B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 1975, 257, 251–253. 10.1038/257251a0. - DOI - PubMed
- Keith J. M.; Ensinger M. J.; Moss B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 1978, 253 (14), 5033–5039. 10.1016/S0021-9258(17)34652-5. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

