DNA corona on nanoparticles leads to an enhanced immunostimulatory effect with implications for autoimmune diseases
- PMID: 38442162
- PMCID: PMC10945806
- DOI: 10.1073/pnas.2319634121
DNA corona on nanoparticles leads to an enhanced immunostimulatory effect with implications for autoimmune diseases
Abstract
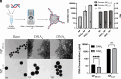
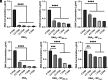
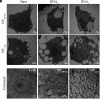
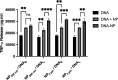
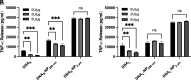
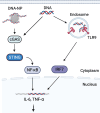
Autoimmune and inflammatory diseases are highly complex, limiting treatment and the development of new therapies. Recent work has shown that cell-free DNA bound to biological microparticles is linked to systemic lupus erythematosus, a prototypic autoimmune disease. However, the heterogeneity and technical challenges associated with the study of biological particles have hindered a mechanistic understanding of their role. Our goal was to develop a well-controlled DNA-particle model system to understand how DNA-particle complexes affect cells. We first characterized the adsorption of DNA on the surface of polystyrene nanoparticles (200 nm and 2 µm) using transmission electron microscopy, dynamic light scattering, and colorimetric DNA concentration assays. We found that DNA adsorbed on the surface of nanoparticles was resistant to degradation by DNase 1. Macrophage cells incubated with the DNA-nanoparticle complexes had increased production of pro-inflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin 6 (IL-6). We probed two intracellular DNA sensing pathways, toll-like receptor 9 (TLR9) and cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING), to determine how cells sense the DNA-nanoparticle complexes. We found that the cGAS-STING pathway is the primary route for the interaction between DNA-nanoparticles and macrophages. These studies provide a molecular and cellular-level understanding of DNA-nanoparticle-macrophage interactions. In addition, this work provides the mechanistic information necessary for future in vivo experiments to elucidate the role of DNA-particle interactions in autoimmune diseases, providing a unique experimental framework to develop novel therapeutic approaches.
Keywords: DNA; autoimmune disease; corona; inflammation; nanoscience.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Aucamp J., Bronkhorst A. J., Badenhorst C. P. S., Pretorius P. J., The diverse origins of circulating cell-free DNA in the human body: A critical re-evaluation of the literature. Biol. Rev. 93, 1649–1683 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

