Rubisco is evolving for improved catalytic efficiency and CO2 assimilation in plants
- PMID: 38442173
- PMCID: PMC10945770
- DOI: 10.1073/pnas.2321050121
Rubisco is evolving for improved catalytic efficiency and CO2 assimilation in plants
Abstract
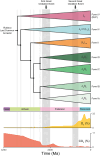
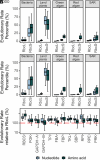
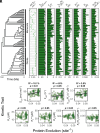
Rubisco is the primary entry point for carbon into the biosphere. However, rubisco is widely regarded as inefficient leading many to question whether the enzyme can adapt to become a better catalyst. Through a phylogenetic investigation of the molecular and kinetic evolution of Form I rubisco we uncover the evolutionary trajectory of rubisco kinetic evolution in angiosperms. We show that rbcL is among the 1% of slowest-evolving genes and enzymes on Earth, accumulating one nucleotide substitution every 0.9 My and one amino acid mutation every 7.2 My. Despite this, rubisco catalysis has been continually evolving toward improved CO2/O2 specificity, carboxylase turnover, and carboxylation efficiency. Consistent with this kinetic adaptation, increased rubisco evolution has led to a concomitant improvement in leaf-level CO2 assimilation. Thus, rubisco has been slowly but continually evolving toward improved catalytic efficiency and CO2 assimilation in plants.
Keywords: adaptation; evolution; kinetics; photosynthesis; rubisco.
Conflict of interest statement
Competing interests statement:S.K. is co-founder of Wild Bioscience Ltd.
Figures


References
-
- Canfield D. E., The early history of atmospheric oxygen: Homage to Robert M. Garrels. Annu. Rev. Earth Planet. Sci. 33, 1–36 (2005).
-
- Kasting J. F., Earth’s early atmosphere. Science 259, 920–926 (1993). - PubMed
-
- Kaufman A. J., et al. , Late archean biospheric oxygenation and atmospheric evolution. Science 317, 1900–1903 (2007). - PubMed
-
- Nisbet E. G., et al. , The age of Rubisco: The evolution of oxygenic photosynthesis. Geobiology 5, 311–335 (2007).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

