eEF1A2 promotes PTEN-GSK3β-SCF complex-dependent degradation of Aurora kinase A and is inactivated in breast cancer
- PMID: 38442201
- PMCID: PMC12039992
- DOI: 10.1126/scisignal.adh4475
eEF1A2 promotes PTEN-GSK3β-SCF complex-dependent degradation of Aurora kinase A and is inactivated in breast cancer
Abstract
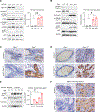
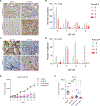
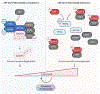
The translation elongation factor eEF1A promotes protein synthesis. Its methylation by METTL13 increases its activity, supporting tumor growth. However, in some cancers, a high abundance of eEF1A isoforms is associated with a good prognosis. Here, we found that eEF1A2 exhibited oncogenic or tumor-suppressor functions depending on its interaction with METTL13 or the phosphatase PTEN, respectively. METTL13 and PTEN competed for interaction with eEF1A2 in the same structural domain. PTEN-bound eEF1A2 promoted the ubiquitination and degradation of the mitosis-promoting Aurora kinase A in the S and G2 phases of the cell cycle. eEF1A2 bridged the interactions between the SKP1-CUL1-FBXW7 (SCF) ubiquitin ligase complex, the kinase GSK3β, and Aurora-A, thereby facilitating the phosphorylation of Aurora-A in a degron site that was recognized by FBXW7. Genetic ablation of Eef1a2 or Pten in mice resulted in a greater abundance of Aurora-A and increased cell cycling in mammary tumors, which was corroborated in breast cancer tissues from patients. Reactivating this pathway using fimepinostat, which relieves inhibitory signaling directed at PTEN and increases FBXW7 expression, combined with inhibiting Aurora-A with alisertib, suppressed breast cancer cell proliferation in culture and tumor growth in vivo. The findings demonstrate a therapeutically exploitable, tumor-suppressive role for eEF1A2 in breast cancer.
Conflict of interest statement
Figures




References
-
- Kulkarni G et al., Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Res Treat 102, 31–41 (2007). - PubMed
-
- Pinke DE, Kalloger SE, Francetic T, Huntsman DG, Lee JM, The prognostic significance of elongation factor eEF1A2 in ovarian cancer. Gynecol Oncol 108, 561–568 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

