Origin, evolution, and maintenance of gene-strand bias in bacteria
- PMID: 38442257
- PMCID: PMC11040001
- DOI: 10.1093/nar/gkae155
Origin, evolution, and maintenance of gene-strand bias in bacteria
Abstract
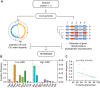
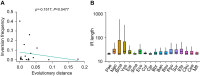
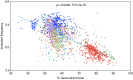
Gene-strand bias is a characteristic feature of bacterial genome organization wherein genes are preferentially encoded on the leading strand of replication, promoting co-orientation of replication and transcription. This co-orientation bias has evolved to protect gene essentiality, expression, and genomic stability from the harmful effects of head-on replication-transcription collisions. However, the origin, variation, and maintenance of gene-strand bias remain elusive. Here, we reveal that the frequency of inversions that alter gene orientation exhibits large variation across bacterial populations and negatively correlates with gene-strand bias. The density, distance, and distribution of inverted repeats show a similar negative relationship with gene-strand bias explaining the heterogeneity in inversions. Importantly, these observations are broadly evident across the entire bacterial kingdom uncovering inversions and inverted repeats as primary factors underlying the variation in gene-strand bias and its maintenance. The distinct catalytic subunits of replicative DNA polymerase have co-evolved with gene-strand bias, suggesting a close link between replication and the origin of gene-strand bias. Congruently, inversion frequencies and inverted repeats vary among bacteria with different DNA polymerases. In summary, we propose that the nature of replication determines the fitness cost of replication-transcription collisions, establishing a selection gradient on gene-strand bias by fine-tuning DNA sequence repeats and, thereby, gene inversions.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






Similar articles
-
A Bacteriophage-Derived Primase-Helicase Orchestrates Plant Organellar DNA Replication.Physiol Plant. 2025 Jul-Aug;177(4):e70379. doi: 10.1111/ppl.70379. Physiol Plant. 2025. PMID: 40620047 Free PMC article.
-
CGGBP1 from higher amniotes restricts cytosine methylation and drives a GC-bias in transcription factor-binding sites at repressed promoters.Transcription. 2025 Jul 31:1-36. doi: 10.1080/21541264.2025.2533598. Online ahead of print. Transcription. 2025. PMID: 40740140
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
PET-CT for assessing mediastinal lymph node involvement in patients with suspected resectable non-small cell lung cancer.Cochrane Database Syst Rev. 2014 Nov 13;2014(11):CD009519. doi: 10.1002/14651858.CD009519.pub2. Cochrane Database Syst Rev. 2014. PMID: 25393718 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
Cited by
-
On the evolution of chromosomal regions with high gene strand bias in bacteria.mBio. 2024 Jun 12;15(6):e0060224. doi: 10.1128/mbio.00602-24. Epub 2024 May 16. mBio. 2024. PMID: 38752745 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

