Homo-BacPROTAC-induced degradation of ClpC1 as a strategy against drug-resistant mycobacteria
- PMID: 38443338
- PMCID: PMC10914731
- DOI: 10.1038/s41467-024-46218-7
Homo-BacPROTAC-induced degradation of ClpC1 as a strategy against drug-resistant mycobacteria
Abstract
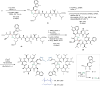
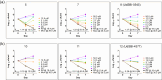
Antimicrobial resistance is a global health threat that requires the development of new treatment concepts. These should not only overcome existing resistance but be designed to slow down the emergence of new resistance mechanisms. Targeted protein degradation, whereby a drug redirects cellular proteolytic machinery towards degrading a specific target, is an emerging concept in drug discovery. We are extending this concept by developing proteolysis targeting chimeras active in bacteria (BacPROTACs) that bind to ClpC1, a component of the mycobacterial protein degradation machinery. The anti-Mycobacterium tuberculosis (Mtb) BacPROTACs are derived from cyclomarins which, when dimerized, generate compounds that recruit and degrade ClpC1. The resulting Homo-BacPROTACs reduce levels of endogenous ClpC1 in Mycobacterium smegmatis and display minimum inhibitory concentrations in the low micro- to nanomolar range in mycobacterial strains, including multiple drug-resistant Mtb isolates. The compounds also kill Mtb residing in macrophages. Thus, Homo-BacPROTACs that degrade ClpC1 represent a different strategy for targeting Mtb and overcoming drug resistance.
© 2024. The Author(s).
Conflict of interest statement
V. M. S., C. K., A. Mantoulidis, P. G., H. W., K. R., K. F., P. B., G. G. and G. B. were employees of Boehringer Ingelheim at the time of this work. The remaining authors declare no competing interests.
Figures




References
-
- Plackett B. No money for new drugs. Nat. Outlook. 2020;586:50–52.
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

