The m6A reader IGF2BP2 regulates glycolytic metabolism and mediates histone lactylation to enhance hepatic stellate cell activation and liver fibrosis
- PMID: 38443347
- PMCID: PMC10914723
- DOI: 10.1038/s41419-024-06509-9
The m6A reader IGF2BP2 regulates glycolytic metabolism and mediates histone lactylation to enhance hepatic stellate cell activation and liver fibrosis
Abstract
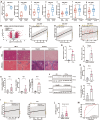
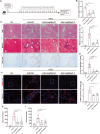
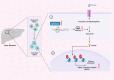
Evidence for the involvement of N6-Methyladenosine (m6A) modification in the etiology and progression of liver fibrosis has emerged and holds promise as a therapeutic target. Insulin-like growth factor 2 (IGF2) mRNA-binding protein 2 (IGF2BP2) is a newly identified m6A-binding protein that functions to enhance mRNA stability and translation. However, its role as an m6A-binding protein in liver fibrosis remains elusive. Here, we observed that IGF2BP2 is highly expressed in liver fibrosis and activated hepatic stellate cells (HSCs), and inhibition of IGF2BP2 protects against HSCs activation and liver fibrogenesis. Mechanistically, as an m6A-binding protein, IGF2BP2 regulates the expression of Aldolase A (ALDOA), a key target in the glycolytic metabolic pathway, which in turn regulates HSCs activation. Furthermore, we observed that active glycolytic metabolism in activated HSCs generates large amounts of lactate as a substrate for histone lactylation. Importantly, histone lactylation transforms the activation phenotype of HSCs. In conclusion, our findings reveal the essential role of IGF2BP2 in liver fibrosis by regulating glycolytic metabolism and highlight the potential of targeting IGF2BP2 as a therapeutic for liver fibrosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Igf2bp2 knockdown improves CCl4-induced liver fibrosis and TGF-β-activated mouse hepatic stellate cells by regulating Tgfbr1.Int Immunopharmacol. 2022 Sep;110:108987. doi: 10.1016/j.intimp.2022.108987. Epub 2022 Jul 9. Int Immunopharmacol. 2022. PMID: 35820364
-
Hexokinase 2-mediated gene expression via histone lactylation is required for hepatic stellate cell activation and liver fibrosis.Cell Metab. 2023 Aug 8;35(8):1406-1423.e8. doi: 10.1016/j.cmet.2023.06.013. Epub 2023 Jul 17. Cell Metab. 2023. PMID: 37463576 Free PMC article.
-
H3K18 lactylation accelerates liver fibrosis progression through facilitating SOX9 transcription.Exp Cell Res. 2024 Jul 15;440(2):114135. doi: 10.1016/j.yexcr.2024.114135. Epub 2024 Jun 18. Exp Cell Res. 2024. PMID: 38901791
-
Epigenetic modifications in hepatic stellate cells contribute to liver fibrosis.Tohoku J Exp Med. 2013 Jan;229(1):35-43. doi: 10.1620/tjem.229.35. Tohoku J Exp Med. 2013. PMID: 23238615 Review.
-
Liver Fibrosis: From Basic Science towards Clinical Progress, Focusing on the Central Role of Hepatic Stellate Cells.Int J Mol Sci. 2024 Jul 18;25(14):7873. doi: 10.3390/ijms25147873. Int J Mol Sci. 2024. PMID: 39063116 Free PMC article. Review.
Cited by
-
New Posttranslational Modification Lactylation Brings New Inspiration for the Treatment of Rheumatoid Arthritis.J Inflamm Res. 2024 Dec 30;17:11845-11860. doi: 10.2147/JIR.S497240. eCollection 2024. J Inflamm Res. 2024. PMID: 39758940 Free PMC article. Review.
-
Targeting Lactic Acid Modification in Ischemic Heart Diseases: Novel Therapeutics and Mechanism.J Cardiovasc Transl Res. 2025 Apr;18(2):257-267. doi: 10.1007/s12265-025-10593-3. Epub 2025 Feb 7. J Cardiovasc Transl Res. 2025. PMID: 39920549 Review.
-
Crosstalk between lactylation and RNA modifications in tumorigenesis: mechanisms and therapeutic implications.Biomark Res. 2025 Aug 26;13(1):110. doi: 10.1186/s40364-025-00824-9. Biomark Res. 2025. PMID: 40859309 Free PMC article. Review.
-
Lactate and lactylation: novel perspectives on fibrosis pathogenesis and therapeutic directions.J Transl Med. 2025 Jul 1;23(1):705. doi: 10.1186/s12967-025-06748-0. J Transl Med. 2025. PMID: 40597258 Free PMC article. Review.
-
Lactylation in health and disease: physiological or pathological?Theranostics. 2025 Jan 2;15(5):1787-1821. doi: 10.7150/thno.105353. eCollection 2025. Theranostics. 2025. PMID: 39897556 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

