Neuronal miR-9 promotes HSV-1 epigenetic silencing and latency by repressing Oct-1 and Onecut family genes
- PMID: 38443365
- PMCID: PMC10914762
- DOI: 10.1038/s41467-024-46057-6
Neuronal miR-9 promotes HSV-1 epigenetic silencing and latency by repressing Oct-1 and Onecut family genes
Abstract
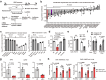
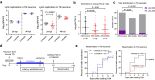
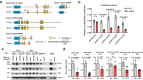
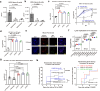
Herpes simplex virus 1 (HSV-1) latent infection entails repression of viral lytic genes in neurons. By functional screening using luciferase-expressing HSV-1, we identify ten neuron-specific microRNAs potentially repressing HSV-1 neuronal replication. Transfection of miR-9, the most active candidate from the screen, decreases HSV-1 replication and gene expression in Neuro-2a cells. Ectopic expression of miR-9 from lentivirus or recombinant HSV-1 suppresses HSV-1 replication in male primary mouse neurons in culture and mouse trigeminal ganglia in vivo, and reactivation from latency in the primary neurons. Target prediction and validation identify transcription factors Oct-1, a known co-activator of HSV transcription, and all three Onecut family members as miR-9 targets. Knockdown of ONECUT2 decreases HSV-1 yields in Neuro-2a cells. Overexpression of each ONECUT protein increases HSV-1 replication in Neuro-2a cells, human induced pluripotent stem cell-derived neurons, and primary mouse neurons, and accelerates reactivation from latency in the mouse neurons. Mutagenesis, ChIP-seq, RNA-seq, ChIP-qPCR and ATAC-seq results suggest that ONECUT2 can nonspecifically bind to viral genes via its CUT domain, globally stimulate viral gene transcription, reduce viral heterochromatin and enhance the accessibility of viral chromatin. Thus, neuronal miR-9 promotes viral epigenetic silencing and latency by targeting multiple host transcription factors important for lytic gene activation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Neuronal miR-138 Represses HSV-2 Lytic Infection by Regulating Viral and Host Genes with Mechanistic Differences from HSV-1.J Virol. 2022 May 11;96(9):e0034922. doi: 10.1128/jvi.00349-22. Epub 2022 Apr 11. J Virol. 2022. PMID: 35404085 Free PMC article.
-
Regulation of host and virus genes by neuronal miR-138 favours herpes simplex virus 1 latency.Nat Microbiol. 2021 May;6(5):682-696. doi: 10.1038/s41564-020-00860-1. Epub 2021 Feb 8. Nat Microbiol. 2021. PMID: 33558653 Free PMC article.
-
Mutations Inactivating Herpes Simplex Virus 1 MicroRNA miR-H2 Do Not Detectably Increase ICP0 Gene Expression in Infected Cultured Cells or Mouse Trigeminal Ganglia.J Virol. 2017 Jan 3;91(2):e02001-16. doi: 10.1128/JVI.02001-16. Print 2017 Jan 15. J Virol. 2017. PMID: 27847363 Free PMC article.
-
A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review.
-
Chromatin-mediated epigenetic regulation of HSV-1 transcription as a potential target in antiviral therapy.Antiviral Res. 2021 Aug;192:105103. doi: 10.1016/j.antiviral.2021.105103. Epub 2021 Jun 1. Antiviral Res. 2021. PMID: 34082058 Free PMC article. Review.
Cited by
-
The opportunities and challenges of epigenetic approaches to manage herpes simplex infections.Expert Rev Anti Infect Ther. 2024 Dec;22(12):1123-1142. doi: 10.1080/14787210.2024.2420329. Epub 2024 Nov 6. Expert Rev Anti Infect Ther. 2024. PMID: 39466139 Review.
-
HSV-1 hijacks mitochondrial dynamics: potential molecular mechanisms linking viral infection to neurodegenerative disorders.Apoptosis. 2025 Jul 1. doi: 10.1007/s10495-025-02142-9. Online ahead of print. Apoptosis. 2025. PMID: 40593393 Review.
-
A review of HSV pathogenesis, vaccine development, and advanced applications.Mol Biomed. 2024 Aug 29;5(1):35. doi: 10.1186/s43556-024-00199-7. Mol Biomed. 2024. PMID: 39207577 Free PMC article. Review.
-
Models of Herpes Simplex Virus Latency.Viruses. 2024 May 8;16(5):747. doi: 10.3390/v16050747. Viruses. 2024. PMID: 38793628 Free PMC article. Review.
-
Global MicroRNA Profiling of HSV-1 Infected Cornea Identifies miR-329 as a Novel Regulator of Virus Infection.Invest Ophthalmol Vis Sci. 2025 Feb 3;66(2):61. doi: 10.1167/iovs.66.2.61. Invest Ophthalmol Vis Sci. 2025. PMID: 39992671 Free PMC article.
References
-
- Knipe, D. M., Heldwein, E. F., Mohr, I. J. & Sodroski, C. N. in Fields Virology Seventh Edition (eds Howley, P. M. et al.) 235–296 (Lippincott Williams & Wilkins, 2021).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

