Genome-wide association analyses of ovarian cancer patients undergoing primary debulking surgery identify candidate genes for residual disease
- PMID: 38443389
- PMCID: PMC10915171
- DOI: 10.1038/s41525-024-00395-y
Genome-wide association analyses of ovarian cancer patients undergoing primary debulking surgery identify candidate genes for residual disease
Abstract
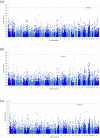
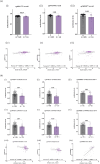
Survival from ovarian cancer depends on the resection status after primary surgery. We performed genome-wide association analyses for resection status of 7705 ovarian cancer patients, including 4954 with high-grade serous carcinoma (HGSOC), to identify variants associated with residual disease. The most significant association with resection status was observed for rs72845444, upstream of MGMT, in HGSOC (p = 3.9 × 10-8). In gene-based analyses, PPP2R5C was the most strongly associated gene in HGSOC after stage adjustment. In an independent set of 378 ovarian tumours from the AGO-OVAR 11 study, variants near MGMT and PPP2R5C correlated with methylation and transcript levels, and PPP2R5C mRNA levels predicted progression-free survival in patients with residual disease. MGMT encodes a DNA repair enzyme, and PPP2R5C encodes the B56γ subunit of the PP2A tumour suppressor. Our results link heritable variation at these two loci with resection status in HGSOC.
© 2024. The Author(s).
Conflict of interest statement
U.C. received honoraria for lectures from Lilly and AstraZeneca and is on the advisory board of AstraZeneca. J.P. received honoraria from Roche Pharma AG, AstraZeneca, Amgen, Clovis Oncology, MSD Oncology, GSK, Chugai Pharma, Teva, Medupdate, SAI MedPartners, Decision Resources, Simon-Kucher and partners, Juniper, Bionest partners, Vox Bio, Axiom healthcare strategies, Prosapient, iMed Institut, Lilly, and is a consultant/advisor for AstraZeneca, Roche, Pharma AG, Tesaro, Clovis Oncology, MSD Oncology. P.A.F. conducts research funded by Amgen, Novartis and Pfizer and received Honoraria from Roche, Novartis and Pfizer. None of these sponsors had any role in the design, data acquisition or interpretation of results in the present study.
Figures


References
Grants and funding
- R21 CA267050/CA/NCI NIH HHS/United States
- K05 CA154337/CA/NCI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- P50 CA105009/CA/NCI NIH HHS/United States
- K07 CA080668/CA/NCI NIH HHS/United States
- P30 CA076292/CA/NCI NIH HHS/United States
- R01 CA076016/CA/NCI NIH HHS/United States
- R01 CA248288/CA/NCI NIH HHS/United States
- U19 CA148112/CA/NCI NIH HHS/United States
- R01 CA149429/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- UL1 TR001881/TR/NCATS NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
- M01 RR000056/RR/NCRR NIH HHS/United States
- R01 CA095023/CA/NCI NIH HHS/United States
- P30 CA047904/CA/NCI NIH HHS/United States
- R01 CA058598/CA/NCI NIH HHS/United States
- R01 CA054419/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

