Improved green and red GRAB sensors for monitoring spatiotemporal serotonin release in vivo
- PMID: 38443508
- PMCID: PMC11377854
- DOI: 10.1038/s41592-024-02188-8
Improved green and red GRAB sensors for monitoring spatiotemporal serotonin release in vivo
Abstract
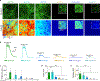
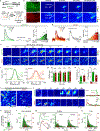
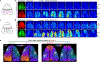
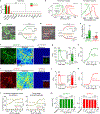
The serotonergic system plays important roles in both physiological and pathological processes, and is a therapeutic target for many psychiatric disorders. Although several genetically encoded GFP-based serotonin (5-HT) sensors were recently developed, their sensitivities and spectral profiles are relatively limited. To overcome these limitations, we optimized green fluorescent G-protein-coupled receptor (GPCR)-activation-based 5-HT (GRAB5-HT) sensors and developed a red fluorescent GRAB5-HT sensor. These sensors exhibit excellent cell surface trafficking and high specificity, sensitivity and spatiotemporal resolution, making them suitable for monitoring 5-HT dynamics in vivo. Besides recording subcortical 5-HT release in freely moving mice, we observed both uniform and gradient 5-HT release in the mouse dorsal cortex with mesoscopic imaging. Finally, we performed dual-color imaging and observed seizure-induced waves of 5-HT release throughout the cortex following calcium and endocannabinoid waves. In summary, these 5-HT sensors can offer valuable insights regarding the serotonergic system in both health and disease.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures











References
-
- Portas CM et al. On-line detection of extracellular levels of serotonin in dorsal raphe nucleus and frontal cortex over the sleep/wake cycle in the freely moving rat. Neuroscience 83, 807–814 (1998). - PubMed
-
- Lesch KP et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274, 1527–1531 (1996). - PubMed
-
- Theodore WH, Juhasz C, Savic V & Drevets W Serotonin, depression, and epilepsy. Epilepsia 46, 3–3 (2005).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

