Multi-ancestry polygenic mechanisms of type 2 diabetes
- PMID: 38443691
- PMCID: PMC11175990
- DOI: 10.1038/s41591-024-02865-3
Multi-ancestry polygenic mechanisms of type 2 diabetes
Erratum in
-
Author Correction: Multi-ancestry polygenic mechanisms of type 2 diabetes.Nat Med. 2024 Jul;30(7):2091. doi: 10.1038/s41591-024-03066-8. Nat Med. 2024. PMID: 38760590 No abstract available.
Abstract
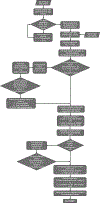
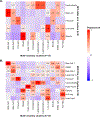
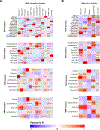
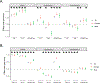
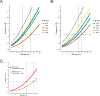
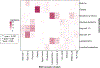
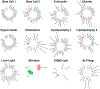
Type 2 diabetes (T2D) is a multifactorial disease with substantial genetic risk, for which the underlying biological mechanisms are not fully understood. In this study, we identified multi-ancestry T2D genetic clusters by analyzing genetic data from diverse populations in 37 published T2D genome-wide association studies representing more than 1.4 million individuals. We implemented soft clustering with 650 T2D-associated genetic variants and 110 T2D-related traits, capturing known and novel T2D clusters with distinct cardiometabolic trait associations across two independent biobanks representing diverse genetic ancestral populations (African, n = 21,906; Admixed American, n = 14,410; East Asian, n =2,422; European, n = 90,093; and South Asian, n = 1,262). The 12 genetic clusters were enriched for specific single-cell regulatory regions. Several of the polygenic scores derived from the clusters differed in distribution among ancestry groups, including a significantly higher proportion of lipodystrophy-related polygenic risk in East Asian ancestry. T2D risk was equivalent at a body mass index (BMI) of 30 kg m-2 in the European subpopulation and 24.2 (22.9-25.5) kg m-2 in the East Asian subpopulation; after adjusting for cluster-specific genetic risk, the equivalent BMI threshold increased to 28.5 (27.1-30.0) kg m-2 in the East Asian group. Thus, these multi-ancestry T2D genetic clusters encompass a broader range of biological mechanisms and provide preliminary insights to explain ancestry-associated differences in T2D risk profiles.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures

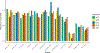




Update of
-
Multi-ancestry Polygenic Mechanisms of Type 2 Diabetes Elucidate Disease Processes and Clinical Heterogeneity.medRxiv [Preprint]. 2023 Sep 29:2023.09.28.23296294. doi: 10.1101/2023.09.28.23296294. medRxiv. 2023. Update in: Nat Med. 2024 Apr;30(4):1065-1074. doi: 10.1038/s41591-024-02865-3. PMID: 37808749 Free PMC article. Updated. Preprint.
-
Multi-ancestry Polygenic Mechanisms of Type 2 Diabetes Elucidate Disease Processes and Clinical Heterogeneity.Res Sq [Preprint]. 2023 Oct 9:rs.3.rs-3399145. doi: 10.21203/rs.3.rs-3399145/v1. Res Sq. 2023. Update in: Nat Med. 2024 Apr;30(4):1065-1074. doi: 10.1038/s41591-024-02865-3. PMID: 37886436 Free PMC article. Updated. Preprint.

