Mesenchymal stem/stromal cells from human pluripotent stem cell-derived brain organoid enhance the ex vivo expansion and maintenance of hematopoietic stem/progenitor cells
- PMID: 38443990
- PMCID: PMC10916050
- DOI: 10.1186/s13287-023-03624-w
Mesenchymal stem/stromal cells from human pluripotent stem cell-derived brain organoid enhance the ex vivo expansion and maintenance of hematopoietic stem/progenitor cells
Abstract
Background: Mesenchymal stem/stromal cells (MSCs) are of great therapeutic value due to their role in maintaining the function of hematopoietic stem/progenitor cells (HSPCs). MSCs derived from human pluripotent stem cells represent an ideal alternative because of their unlimited supply. However, the role of MSCs with neural crest origin derived from HPSCs on the maintenance of HSPCs has not been reported.
Methods: Flow cytometric analysis, RNA sequencing and differentiation ability were applied to detect the characteristics of stromal cells from 3D human brain organoids. Human umbilical cord blood CD34+ (UCB-CD34+) cells were cultured in different coculture conditions composed of stromal cells and umbilical cord MSCs (UC-MSCs) with or without a cytokine cocktail. The hematopoietic stroma capacity of stromal cells was tested in vitro with the LTC-IC assay and in vivo by cotransplantation of cord blood nucleated cells and stroma cells into immunodeficient mice. RNA and proteomic sequencing were used to detect the role of MSCs on HSPCs.
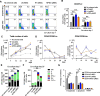
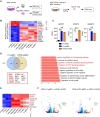
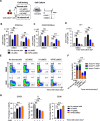
Results: The stromal cells, derived from both H1-hESCs and human induced pluripotent stem cells forebrain organoids, were capable of differentiating into the classical mesenchymal-derived cells (osteoblasts, chondrocytes, and adipocytes). These cells expressed MSC markers, thus named pluripotent stem cell-derived MSCs (pMSCs). The pMSCs showed neural crest origin with CD271 expression in the early stage. When human UCB-CD34+ HSPCs were cocultured on UC-MSCs or pMSCs, the latter resulted in robust expansion of UCB-CD34+ HSPCs in long-term culture and efficient maintenance of their transplantability. Comparison by RNA sequencing indicated that coculture of human UCB-CD34+ HSPCs with pMSCs provided an improved microenvironment for HSC maintenance. The pMSCs highly expressed the Wnt signaling inhibitors SFRP1 and SFRP2, indicating that they may help to modulate the cell cycle to promote the maintenance of UCB-CD34+ HSPCs by antagonizing Wnt activation.
Conclusions: A novel method for harvesting MSCs with neural crest origin from 3D human brain organoids under serum-free culture conditions was reported. We demonstrate that the pMSCs support human UCB-HSPC expansion in vitro in a long-term culture and the maintenance of their transplantable ability. RNA and proteomic sequencing indicated that pMSCs provided an improved microenvironment for HSC maintenance via mechanisms involving cell-cell contact and secreted factors and suppression of Wnt signaling. This represents a novel method for large-scale production of MSCs of neural crest origin and provides a potential approach for development of human hematopoietic stromal cell therapy for treatment of dyshematopoiesis.
Keywords: HSC transplantation; HSCs; Human brain organoids; MSCs; Wnt signaling pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Oostendorp RA, Harvey KN, Kusadasi N, de Bruijn MF, Saris C, Ploemacher RE, Medvinsky AL, Dzierzak EA. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood. 2002;99(4):1183–1189. doi: 10.1182/blood.V99.4.1183. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

