GhCTSF1, a short PPR protein with a conserved role in chloroplast development and photosynthesis, participates in intron splicing of rpoC1 and ycf3-2 transcripts in cotton
- PMID: 38444162
- PMCID: PMC11211521
- DOI: 10.1016/j.xplc.2024.100858
GhCTSF1, a short PPR protein with a conserved role in chloroplast development and photosynthesis, participates in intron splicing of rpoC1 and ycf3-2 transcripts in cotton
Abstract
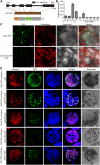
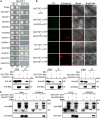
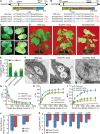
Cotton is one of the most important textile fibers worldwide. As crucial agronomic traits, leaves play an essential role in the growth, disease resistance, fiber quality, and yield of cotton plants. Pentatricopeptide repeat (PPR) proteins are a large family of nuclear-encoded proteins involved in organellar or nuclear RNA metabolism. Using a virus-induced gene silencing assay, we found that cotton plants displayed variegated yellow leaf phenotypes with decreased chlorophyll content when expression of the PPR gene GhCTSF1 was silenced. GhCTSF1 encodes a chloroplast-localized protein that contains only two PPR motifs. Disruption of GhCTSF1 substantially reduces the splicing efficiency of rpoC1 intron 1 and ycf3 intron 2. Loss of function of the GhCTSF1 ortholog EMB1417 causes splicing defects in rpoC1 and ycf3-2, leading to impaired chloroplast structure and decreased photosynthetic rates in Arabidopsis. We also found that GhCTSF1 interacts with two splicing factors, GhCRS2 and GhWTF1. Defects in GhCRS2 and GhWTF1 severely affect intron splicing of rpoC1 and ycf3-2 in cotton, leading to defects in chloroplast development and a reduction in photosynthesis. Our results suggest that GhCTSF1 is specifically required for splicing rpoC1 and ycf3-2 in cooperation with GhCRS2 and GhWTF1.
Keywords: PPR protein; RNA splicing; chloroplast; cotton; photosynthesis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
GhCTEF2 encodes a PLS-type PPR protein required for chloroplast development and plastid RNA editing in cotton.Plant Sci. 2025 Jun;355:112478. doi: 10.1016/j.plantsci.2025.112478. Epub 2025 Mar 17. Plant Sci. 2025. PMID: 40107517
-
OsPPR11 encoding P-type PPR protein that affects group II intron splicing and chloroplast development.Plant Cell Rep. 2023 Feb;42(2):421-431. doi: 10.1007/s00299-022-02968-6. Epub 2022 Dec 28. Plant Cell Rep. 2023. PMID: 36576552
-
P-Type Pentatricopeptide Repeat Proteins YS1 and YS2 Function in Splicing of petB Intron to Maintain Chloroplast Homeostasis During Rice Seedling Development.Int J Mol Sci. 2025 May 7;26(9):4459. doi: 10.3390/ijms26094459. Int J Mol Sci. 2025. PMID: 40362697 Free PMC article.
-
Functioning of PPR Proteins in Organelle RNA Metabolism and Chloroplast Biogenesis.Front Plant Sci. 2021 Feb 9;12:627501. doi: 10.3389/fpls.2021.627501. eCollection 2021. Front Plant Sci. 2021. PMID: 33633768 Free PMC article. Review.
-
PPR proteins - orchestrators of organelle RNA metabolism.Physiol Plant. 2019 May;166(1):451-459. doi: 10.1111/ppl.12950. Physiol Plant. 2019. PMID: 30809817 Review.
Cited by
-
Pentatricopeptide repeat proteins in plants: Cellular functions, action mechanisms, and potential applications.Plant Commun. 2025 Feb 10;6(2):101203. doi: 10.1016/j.xplc.2024.101203. Epub 2024 Dec 5. Plant Commun. 2025. PMID: 39644091 Free PMC article. Review.
References
-
- Barkan A., Small I. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 2014;65:415–442. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

