More than the sum of its parts: uncovering emerging effects of microbial interactions in complex communities
- PMID: 38444203
- PMCID: PMC10950044
- DOI: 10.1093/femsec/fiae029
More than the sum of its parts: uncovering emerging effects of microbial interactions in complex communities
Abstract
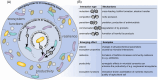
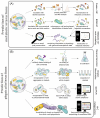
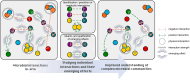
Microbial communities are not only shaped by the diversity of microorganisms and their individual metabolic potential, but also by the vast amount of intra- and interspecies interactions that can occur pairwise interactions among microorganisms, we suggest that more attention should be drawn towards the effects on the entire microbiome that emerge from individual interactions between community members. The production of certain metabolites that can be tied to a specific microbe-microbe interaction might subsequently influence the physicochemical parameters of the habitat, stimulate a change in the trophic network of the community or create new micro-habitats through the formation of biofilms, similar to the production of antimicrobial substances which might negatively affect only one microorganism but cause a ripple effect on the abundance of other community members. Here, we argue that combining established as well as innovative laboratory and computational methods is needed to predict novel interactions and assess their secondary effects. Such efforts will enable future microbiome studies to expand our knowledge on the dynamics of complex microbial communities.
Keywords: community dynamics; microbial interactions; microbiome research; secondary effects.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare no competing interests.
Figures