N-glycosylation of the SARS-CoV-2 spike protein at Asn331 and Asn343 is involved in spike-ACE2 binding, virus entry, and regulation of IL-6
- PMID: 38444370
- PMCID: PMC11273356
- DOI: 10.1111/1348-0421.13121
N-glycosylation of the SARS-CoV-2 spike protein at Asn331 and Asn343 is involved in spike-ACE2 binding, virus entry, and regulation of IL-6
Abstract
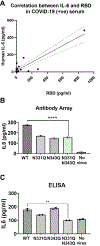
The coronavirus disease 2019 (COVID-19) pandemic is an ongoing global public health crisis. The causative agent, the SARS-CoV-2 virus, enters host cells via molecular interactions between the viral spike protein and the host cell ACE2 surface protein. The SARS-CoV-2 spike protein is extensively decorated with up to 66 N-linked glycans. Glycosylation of viral proteins is known to function in immune evasion strategies but may also function in the molecular events of viral entry into host cells. Here, we show that N-glycosylation at Asn331 and Asn343 of SARS-CoV-2 spike protein is required for it to bind to ACE2 and for the entry of pseudovirus harboring the SARS-CoV-2 spike protein into cells. Interestingly, high-content glycan binding screening data have shown that N-glycosylation of Asn331 and Asn343 of the RBD is important for binding to the specific glycan molecule G4GN (Galβ-1,4 GlcNAc), which is critical for spike-RBD-ACE2 binding. Furthermore, IL-6 was identified through antibody array analysis of conditioned media of the corresponding pseudovirus assay. Mutation of N-glycosylation of Asn331 and Asn343 sites of the spike receptor-binding domain (RBD) significantly reduced the transcriptional upregulation of pro-inflammatory signaling molecule IL-6. In addition, IL-6 levels correlated with spike protein levels in COVID-19 patients' serum. These findings establish the importance of RBD glycosylation in SARS-CoV-2 pathogenesis, which can be exploited for the development of novel therapeutics for COVID-19.
Keywords: ACE2; COVID‐19; IL‐6; N‐glycosylation; SARS‐CoV‐2; receptor‐binding domain; spike protein.
© 2024 The Authors. Microbiology and Immunology published by The Societies and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
Figures



Similar articles
-
Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration.Elife. 2020 Oct 26;9:e61552. doi: 10.7554/eLife.61552. Elife. 2020. PMID: 33103998 Free PMC article.
-
Characterization of Site-Specific N- and O-Glycopeptides from Recombinant Spike and ACE2 Glycoproteins Using LC-MS/MS Analysis.Int J Mol Sci. 2024 Dec 20;25(24):13649. doi: 10.3390/ijms252413649. Int J Mol Sci. 2024. PMID: 39769415 Free PMC article.
-
SARS-CoV-2 spike protein receptor-binding domain N-glycans facilitate viral internalization in respiratory epithelial cells.Biochem Biophys Res Commun. 2021 Nov 19;579:69-75. doi: 10.1016/j.bbrc.2021.09.053. Epub 2021 Sep 23. Biochem Biophys Res Commun. 2021. PMID: 34592572 Free PMC article.
-
A Deadly Embrace: Hemagglutination Mediated by SARS-CoV-2 Spike Protein at Its 22 N-Glycosylation Sites, Red Blood Cell Surface Sialoglycoproteins, and Antibody.Int J Mol Sci. 2022 Feb 25;23(5):2558. doi: 10.3390/ijms23052558. Int J Mol Sci. 2022. PMID: 35269703 Free PMC article. Review.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
Cited by
-
Bioinformatic Selection of Mannose-Specific Lectins from Allium genus as SARS-CoV-2 Inhibitors Analysing Protein-Protein Interaction.Life (Basel). 2025 Jan 23;15(2):162. doi: 10.3390/life15020162. Life (Basel). 2025. PMID: 40003571 Free PMC article.
References
-
- Chapter Payne S. 17 - Family Coronaviridae. In: Payne S, editor. Viruses: From Understanding to Investigation: Academic Press; 2017. p. 149–58.
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1953–66. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

