METTL3 as a master regulator of translation in cancer: mechanisms and implications
- PMID: 38444581
- PMCID: PMC10914372
- DOI: 10.1093/narcan/zcae009
METTL3 as a master regulator of translation in cancer: mechanisms and implications
Abstract
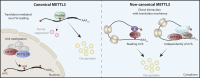
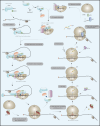
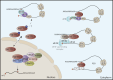
Translational regulation is an important step in the control of gene expression. In cancer cells, the orchestration of both global control of protein synthesis and selective translation of specific mRNAs promote tumor cell survival, angiogenesis, transformation, invasion and metastasis. N6-methyladenosine (m6A), the most prevalent mRNA modification in higher eukaryotes, impacts protein translation. Over the past decade, the development of m6A mapping tools has facilitated comprehensive functional investigations, revealing the involvement of this chemical mark, together with its writer METTL3, in promoting the translation of both oncogenes and tumor suppressor transcripts, with the impact being context-dependent. This review aims to consolidate our current understanding of how m6A and METTL3 shape translation regulation in the realm of cancer biology. In addition, it delves into the role of cytoplasmic METTL3 in protein synthesis, operating independently of its catalytic activity. Ultimately, our goal is to provide critical insights into the interplay between m6A, METTL3 and translational regulation in cancer, offering a deeper comprehension of the mechanisms sustaining tumorigenesis.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Cancer.
Figures



References
-
- Scherrer K. Primary transcripts: from the discovery of RNA processing to current concepts of gene expression - review. Exp. Cell. Res. 2018; 373:1–33. - PubMed
-
- Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999; 98:1–4. - PubMed
-
- Silvera D., Formenti S.C., Schneider R.J. Translational control in cancer. Nat. Rev. Cancer. 2010; 10:254–266. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

