Prdm15 acts upstream of Wnt4 signaling in anterior neural development of Xenopus laevis
- PMID: 38444828
- PMCID: PMC10912572
- DOI: 10.3389/fcell.2024.1316048
Prdm15 acts upstream of Wnt4 signaling in anterior neural development of Xenopus laevis
Abstract
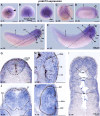
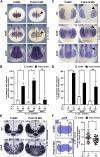
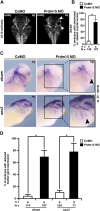
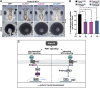
Mutations in PRDM15 lead to a syndromic form of holoprosencephaly (HPE) known as the Galloway-Mowat syndrome (GAMOS). While a connection between PRDM15, a zinc finger transcription factor, and WNT/PCP signaling has been established, there is a critical need to delve deeper into their contributions to early development and GAMOS pathogenesis. We used the South African clawed frog Xenopus laevis as the vertebrate model organism and observed that prdm15 was enriched in the tissues and organs affected in GAMOS. Furthermore, we generated a morpholino oligonucleotide-mediated prdm15 knockdown model showing that the depletion of Prdm15 leads to abnormal eye, head, and brain development, effectively recapitulating the anterior neural features in GAMOS. An analysis of the underlying molecular basis revealed a reduced expression of key genes associated with eye, head, and brain development. Notably, this reduction could be rescued by the introduction of wnt4 RNA, particularly during the induction of the respective tissues. Mechanistically, our data demonstrate that Prdm15 acts upstream of both canonical and non-canonical Wnt4 signaling during anterior neural development. Our findings describe severe ocular and anterior neural abnormalities upon Prdm15 depletion and elucidate the role of Prdm15 in canonical and non-canonical Wnt4 signaling.
Keywords: GAMOS; HPE; Prdm15; Wnt signaling; Xenopus laevis; disease modeling.
Copyright © 2024 Saumweber, Mzoughi, Khadra, Werberger, Schumann, Guccione, Schmeisser and Kühl.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors have declared that they were an editorial board member of Frontiers at the time of submission. This had no impact on the peer review process and the final decision.
Figures





Similar articles
-
Mutations in PRDM15 Are a Novel Cause of Galloway-Mowat Syndrome.J Am Soc Nephrol. 2021 Mar;32(3):580-596. doi: 10.1681/ASN.2020040490. Epub 2021 Feb 16. J Am Soc Nephrol. 2021. PMID: 33593823 Free PMC article.
-
PRDM15 loss of function links NOTCH and WNT/PCP signaling to patterning defects in holoprosencephaly.Sci Adv. 2020 Jan 10;6(2):eaax9852. doi: 10.1126/sciadv.aax9852. eCollection 2020 Jan. Sci Adv. 2020. PMID: 31950080 Free PMC article.
-
Galloway-Mowat syndrome: New insights from bioinformatics and expression during Xenopus embryogenesis.Gene Expr Patterns. 2021 Dec;42:119215. doi: 10.1016/j.gep.2021.119215. Epub 2021 Oct 4. Gene Expr Patterns. 2021. PMID: 34619372
-
Amphibian Zic Genes.Adv Exp Med Biol. 2018;1046:107-140. doi: 10.1007/978-981-10-7311-3_7. Adv Exp Med Biol. 2018. PMID: 29442320 Review.
-
Novel homozygous OSGEP gene pathogenic variants in two unrelated patients with Galloway-Mowat syndrome: case report and review of the literature.BMC Nephrol. 2019 Apr 11;20(1):126. doi: 10.1186/s12882-019-1317-y. BMC Nephrol. 2019. PMID: 30975089 Free PMC article. Review.
Cited by
-
Molecular evolution of the transcription factor PRDM genes and expression profiles in response to stimulations and spinal cord injury repair in lamprey (Lethenteron reissneri).Immunogenetics. 2025 Jul 14;77(1):25. doi: 10.1007/s00251-025-01382-y. Immunogenetics. 2025. PMID: 40658216
-
The Role of Purine Metabolism and Uric Acid in Postnatal Neurologic Development.Molecules. 2025 Feb 11;30(4):839. doi: 10.3390/molecules30040839. Molecules. 2025. PMID: 40005150 Free PMC article. Review.
-
Comparing the effects of three neonicotinoids on embryogenesis of the South African clawed frog Xenopus laevis.Curr Res Toxicol. 2024 Apr 21;6:100169. doi: 10.1016/j.crtox.2024.100169. eCollection 2024. Curr Res Toxicol. 2024. PMID: 38706785 Free PMC article.
References
LinkOut - more resources
Full Text Sources

