Elotuzumab Enhances CD16-Independent NK Cell-Mediated Cytotoxicity against Myeloma Cells by Upregulating Several NK Cell-Enhancing Genes
- PMID: 38444839
- PMCID: PMC10914431
- DOI: 10.1155/2024/1429879
Elotuzumab Enhances CD16-Independent NK Cell-Mediated Cytotoxicity against Myeloma Cells by Upregulating Several NK Cell-Enhancing Genes
Abstract
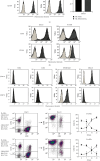
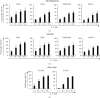
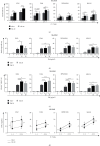
Multiple myeloma (MM) is an intractable hematological malignancy caused by abnormalities in plasma cells. Combination therapy using antibodies and natural killer (NK) effectors, which are innate immune cells with safe and potent antitumor activity, is a promising approach for cancer immunotherapy and can enhance antitumor effects. Elotuzumab (Elo) is an immune-stimulatory antibody that targets the signaling lymphocytic activation molecule family 7 (SLAMF7) expressed on the surface of MM and NK cells. We confirmed that Elo strongly promoted NK cell-mediated antibody-dependent cellular cytotoxicity (ADCC) against SLAMF7-positive MM cells in a CD16-dependent NK cell line, and also activated expanded NK cells derived from peripheral blood mononuclear cells of healthy donors and patients with MM in the present study. However, the antitumor effects and genes involved in the direct promotion of NK cell-mediated activation using Elo in CD16-independent NK cells are not clearly known. In this study, we demonstrated that Elo pretreatment significantly enhanced CD16-independent NK cell-mediated cytotoxicity in both SLAMF7-positive MM.1S and SLAMF7-negative K562, U266, and RPMI 8226 tumor cells. Upon direct simulation of CD16-independent NK cells with Elo, increased levels of CD107a degranulation and IFN-γ secretion were observed along with the upregulation of granzyme B, TNF-α, and IL-1α gene expression. The enhanced NK cell function could also be attributed to the increased expression of the transcription factors T-BET and EOMES. Furthermore, the augmentation of the antitumor effects of CD16-independent NK cells upon pretreatment with Elo enhanced the expression of CRTAM, TNFRSF9, EAT-2, and FOXP3 genes and reduced the expression of HSPA6. Our results suggest that Elo directly promotes the cytotoxic function of CD16-independent NK cells against target cells, which is associated with the upregulation of the expression of several NK cell-enhancing genes.
Copyright © 2024 Yan-Hua Wang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



Similar articles
-
Enhanced SLAMF7 Homotypic Interactions by Elotuzumab Improves NK Cell Killing of Multiple Myeloma.Cancer Immunol Res. 2019 Oct;7(10):1633-1646. doi: 10.1158/2326-6066.CIR-18-0579. Epub 2019 Aug 20. Cancer Immunol Res. 2019. PMID: 31431433 Free PMC article.
-
Elotuzumab, a potential therapeutic humanized anti-SLAMF7 monoclonal antibody, enhances natural killer cell-mediated killing of primary effusion lymphoma cells.Cancer Immunol Immunother. 2022 Oct;71(10):2497-2509. doi: 10.1007/s00262-022-03177-6. Epub 2022 Mar 9. Cancer Immunol Immunother. 2022. PMID: 35262781 Free PMC article.
-
Mechanisms of NK Cell Activation and Clinical Activity of the Therapeutic SLAMF7 Antibody, Elotuzumab in Multiple Myeloma.Front Immunol. 2018 Nov 5;9:2551. doi: 10.3389/fimmu.2018.02551. eCollection 2018. Front Immunol. 2018. PMID: 30455698 Free PMC article. Review.
-
Elotuzumab enhances natural killer cell activation and myeloma cell killing through interleukin-2 and TNF-α pathways.Cancer Immunol Immunother. 2015 Jan;64(1):61-73. doi: 10.1007/s00262-014-1610-3. Epub 2014 Oct 7. Cancer Immunol Immunother. 2015. PMID: 25287778 Free PMC article.
-
Treatment of multiple myeloma with the immunostimulatory SLAMF7 antibody elotuzumab.Ther Adv Hematol. 2016 Oct;7(5):288-301. doi: 10.1177/2040620716657993. Epub 2016 Jul 15. Ther Adv Hematol. 2016. PMID: 27695618 Free PMC article. Review.
Cited by
-
Avelumab induces greater Fc-Fc receptor-dependent natural killer cell activation and dendritic cell crosstalk compared to durvalumab.Oncoimmunology. 2025 Dec;14(1):2494995. doi: 10.1080/2162402X.2025.2494995. Epub 2025 May 1. Oncoimmunology. 2025. PMID: 40311014 Free PMC article.
-
Recent advances in and applications of ex vivo drug sensitivity analysis for blood cancers.Blood Res. 2024 Nov 6;59(1):37. doi: 10.1007/s44313-024-00032-8. Blood Res. 2024. PMID: 39503808 Free PMC article. Review.
-
The Genetic and Molecular Drivers of Multiple Myeloma: Current Insights, Clinical Implications, and the Path Forward.Pharmgenomics Pers Med. 2024 Dec 21;17:573-609. doi: 10.2147/PGPM.S350238. eCollection 2024. Pharmgenomics Pers Med. 2024. PMID: 39723112 Free PMC article. Review.
-
Advancements in adoptive CAR immune cell immunotherapy synergistically combined with multimodal approaches for tumor treatment.Bioact Mater. 2024 Sep 10;42:379-403. doi: 10.1016/j.bioactmat.2024.08.046. eCollection 2024 Dec. Bioact Mater. 2024. PMID: 39308543 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

