Immunological signatures unveiled by integrative systems vaccinology characterization of dengue vaccination trials and natural infection
- PMID: 38444851
- PMCID: PMC10912564
- DOI: 10.3389/fimmu.2024.1282754
Immunological signatures unveiled by integrative systems vaccinology characterization of dengue vaccination trials and natural infection
Abstract
Introduction: Dengue virus infection is a global health problem lacking specific therapy, requiring an improved understanding of DENV immunity and vaccine responses. Considering the recent emerging of new dengue vaccines, here we performed an integrative systems vaccinology characterization of molecular signatures triggered by the natural DENV infection (NDI) and attenuated dengue virus infection models (DVTs).
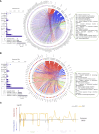
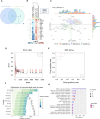
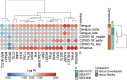
Methods and results: We analyzed 955 samples of transcriptomic datasets of patients with NDI and attenuated dengue virus infection trials (DVT1, DVT2, and DVT3) using a systems vaccinology approach. Differential expression analysis identified 237 common differentially expressed genes (DEGs) between DVTs and NDI. Among them, 28 and 60 DEGs were up or downregulated by dengue vaccination during DVT2 and DVT3, respectively, with 20 DEGs intersecting across all three DVTs. Enriched biological processes of these genes included type I/II interferon signaling, cytokine regulation, apoptosis, and T-cell differentiation. Principal component analysis based on 20 common DEGs (overlapping between DVTs and our NDI validation dataset) distinguished dengue patients by disease severity, particularly in the late acute phase. Machine learning analysis ranked the ten most critical predictors of disease severity in NDI, crucial for the anti-viral immune response.
Conclusion: This work provides insights into the NDI and vaccine-induced overlapping immune response and suggests molecular markers (e.g., IFIT5, ISG15, and HERC5) for anti-dengue-specific therapies and effective vaccination development.
Keywords: dengue; immune response; systems vaccinology; transcriptional signature; vaccine.
Copyright © 2024 Plaça, Fonseca, Marques, Zaki Pour, Usuda, Baiocchi, Prado, Salgado, Filgueiras, Freire, Rocha, Camara, Catar, Moll, Jurisica, Calich, Giil, Rivino, Ochs, Cabral-Miranda, Schimke and Cabral-Marques.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. This study received funding from IBM. The funder had the following involvement in the study: production of figures, revision, and manuscript editing. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures






Similar articles
-
Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus.Sci Rep. 2021 Oct 5;11(1):19707. doi: 10.1038/s41598-021-99227-7. Sci Rep. 2021. PMID: 34611250 Free PMC article.
-
Early Transcriptional Signatures of the Immune Response to a Live Attenuated Tetravalent Dengue Vaccine Candidate in Non-human Primates.PLoS Negl Trop Dis. 2016 May 23;10(5):e0004731. doi: 10.1371/journal.pntd.0004731. eCollection 2016 May. PLoS Negl Trop Dis. 2016. PMID: 27214236 Free PMC article.
-
Human CD4+ T Cell Responses to an Attenuated Tetravalent Dengue Vaccine Parallel Those Induced by Natural Infection in Magnitude, HLA Restriction, and Antigen Specificity.J Virol. 2017 Feb 14;91(5):e02147-16. doi: 10.1128/JVI.02147-16. Print 2017 Mar 1. J Virol. 2017. PMID: 27974563 Free PMC article.
-
Which Dengue Vaccine Approach Is the Most Promising, and Should We Be Concerned about Enhanced Disease after Vaccination? The Path to a Dengue Vaccine: Learning from Human Natural Dengue Infection Studies and Vaccine Trials.Cold Spring Harb Perspect Biol. 2018 Jun 1;10(6):a029371. doi: 10.1101/cshperspect.a029371. Cold Spring Harb Perspect Biol. 2018. PMID: 28716891 Free PMC article. Review.
-
Dengue: Status of current and under-development vaccines.Rev Med Virol. 2020 Jul;30(4):e2101. doi: 10.1002/rmv.2101. Epub 2020 Feb 26. Rev Med Virol. 2020. PMID: 32101634 Review.
Cited by
-
Immunologic Crosstalk and Host-Specific Immune Signature Associated with Dengue.ACS Omega. 2024 Aug 20;9(36):37418-37429. doi: 10.1021/acsomega.4c02506. eCollection 2024 Sep 10. ACS Omega. 2024. PMID: 39281909 Free PMC article. Review.
References
-
- World Health Organization . Dengue and Severe Dengue: WHO; (2015), online. Available at: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

