Microbiome-based classification models for fresh produce safety and quality evaluation
- PMID: 38445872
- PMCID: PMC10986475
- DOI: 10.1128/spectrum.03448-23
Microbiome-based classification models for fresh produce safety and quality evaluation
Abstract
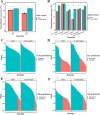
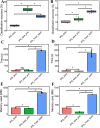
Small sample sizes and loss of sequencing reads during the microbiome data preprocessing can limit the statistical power of differentiating fresh produce phenotypes and prevent the detection of important bacterial species associated with produce contamination or quality reduction. Here, we explored a machine learning-based k-mer hash analysis strategy to identify DNA signatures predictive of produce safety (PS) and produce quality (PQ) and compared it against the amplicon sequence variant (ASV) strategy that uses a typical denoising step and ASV-based taxonomy strategy. Random forest-based classifiers for PS and PQ using 7-mer hash data sets had significantly higher classification accuracy than those using the ASV data sets. We also demonstrated that the proposed combination of integrating multiple data sets and leveraging a 7-mer hash strategy leads to better classification performance for PS and PQ compared to the ASV method but presents lower PS classification accuracy compared to the feature-selected ASV-based taxonomy strategy. Due to the current limitation of generating taxonomy using the 7-mer hash strategy, the ASV-based taxonomy strategy with remarkably less computing time and memory usage is more efficient for PS and PQ classification and applicable for important taxa identification. Results generated from this study lay the foundation for future studies that wish and need to incorporate and/or compare different microbiome sequencing data sets for the application of machine learning in the area of microbial safety and quality of food.
Importance: Identification of generalizable indicators for produce safety (PS) and produce quality (PQ) improves the detection of produce contamination and quality decline. However, effective sequencing read loss during microbiome data preprocessing and the limited sample size of individual studies restrain statistical power to identify important features contributing to differentiating PS and PQ phenotypes. We applied machine learning-based models using individual and integrated k-mer hash and amplicon sequence variant (ASV) data sets for PS and PQ classification and evaluated their classification performance and found that random forest (RF)-based models using integrated 7-mer hash data sets achieved significantly higher PS and PQ classification accuracy. Due to the limitation of taxonomic analysis for the 7-mer hash, we also developed RF-based models using feature-selected ASV-based taxonomic data sets, which performed better PS classification than those using the integrated 7-mer hash data set. The RF feature selection method identified 480 PS indicators and 263 PQ indicators with a positive contribution to the PS and PQ classification.
Keywords: amplicon sequence variant; k-mer hash; machine learning; produce quality; produce safety; random forest.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Jackson C, Stone B, Tyler H. 2015. Emerging perspectives on the natural microbiome of fresh produce vegetables. Agriculture 5:170–187. doi: 10.3390/agriculture5020170 - DOI
-
- Ceuppens S, Delbeke S, De Coninck D, Boussemaere J, Boon N, Uyttendaele M. 2015. Characterization of the bacterial community naturally present on commercially grown basil leaves: evaluation of sample preparation prior to culture-independent techniques. Int J Environ Res Public Health 12:10171–10197. doi: 10.3390/ijerph120810171 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

