Phylogenomics reveals extensive misidentification of fungal strains from the genus Aspergillus
- PMID: 38445873
- PMCID: PMC10986620
- DOI: 10.1128/spectrum.03980-23
Phylogenomics reveals extensive misidentification of fungal strains from the genus Aspergillus
Abstract
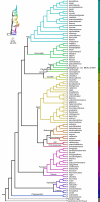
Modern taxonomic classification is often based on phylogenetic analyses of a few molecular markers, although single-gene studies are still common. Here, we leverage genome-scale molecular phylogenetics (phylogenomics) of species and populations to reconstruct evolutionary relationships in a dense data set of 710 fungal genomes from the biomedically and technologically important genus Aspergillus. To do so, we generated a novel set of 1,362 high-quality molecular markers specific for Aspergillus and provided profile Hidden Markov Models for each, facilitating their use by others. Examining the resulting phylogeny helped resolve ongoing taxonomic controversies, identified new ones, and revealed extensive strain misidentification (7.59% of strains were previously misidentified), underscoring the importance of population-level sampling in species classification. These findings were corroborated using the current standard, taxonomically informative loci. These findings suggest that phylogenomics of species and populations can facilitate accurate taxonomic classifications and reconstructions of the Tree of Life.IMPORTANCEIdentification of fungal species relies on the use of molecular markers. Advances in genomic technologies have made it possible to sequence the genome of any fungal strain, making it possible to use genomic data for the accurate assignment of strains to fungal species (and for the discovery of new ones). We examined the usefulness and current limitations of genomic data using a large data set of 710 publicly available genomes from multiple strains and species of the biomedically, agriculturally, and industrially important genus Aspergillus. Our evolutionary genomic analyses revealed that nearly 8% of publicly available Aspergillus genomes are misidentified. Our work highlights the usefulness of genomic data for fungal systematic biology and suggests that systematic genome sequencing of multiple strains, including reference strains (e.g., type strains), of fungal species will be required to reduce misidentification errors in public databases.
Keywords: Aspergillaceae; Penicillium; genomics; pathogen; pathogenicity; plant pathogen; taxogenomics; taxonomy; virulence.
Conflict of interest statement
J.L.S. is an advisor for ForensisGroup Incorporated. A.R. is a scientific consultant for LifeMine Therapeutics, Inc.
Figures


Similar articles
-
A Robust Phylogenomic Time Tree for Biotechnologically and Medically Important Fungi in the Genera Aspergillus and Penicillium.mBio. 2019 Jul 9;10(4):e00925-19. doi: 10.1128/mBio.00925-19. mBio. 2019. PMID: 31289177 Free PMC article.
-
Modern taxonomy of biotechnologically important Aspergillus and Penicillium species.Adv Appl Microbiol. 2014;86:199-249. doi: 10.1016/B978-0-12-800262-9.00004-4. Adv Appl Microbiol. 2014. PMID: 24377856 Review.
-
An evolutionary genomic approach reveals both conserved and species-specific genetic elements related to human disease in closely related Aspergillus fungi.Genetics. 2021 Jun 24;218(2):iyab066. doi: 10.1093/genetics/iyab066. Genetics. 2021. PMID: 33944921 Free PMC article.
-
Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus.Genome Biol. 2017 Feb 14;18(1):28. doi: 10.1186/s13059-017-1151-0. Genome Biol. 2017. PMID: 28196534 Free PMC article.
-
Genomics of the fungal kingdom: insights into eukaryotic biology.Genome Res. 2005 Dec;15(12):1620-31. doi: 10.1101/gr.3767105. Genome Res. 2005. PMID: 16339359 Review.
Cited by
-
Genome-wide patterns of non-coding sequence variation in the major fungal pathogen Aspergillus fumigatus.bioRxiv [Preprint]. 2024 Jan 9:2024.01.08.574724. doi: 10.1101/2024.01.08.574724. bioRxiv. 2024. Update in: G3 (Bethesda). 2024 Jul 8;14(7):jkae091. doi: 10.1093/g3journal/jkae091. PMID: 38260267 Free PMC article. Updated. Preprint.
-
Additions to Pleosporalean Taxa Associated with Xanthoceras sorbifolium from Jilin and Hebei, China.Microorganisms. 2025 May 31;13(6):1296. doi: 10.3390/microorganisms13061296. Microorganisms. 2025. PMID: 40572184 Free PMC article.
-
Overview and Diversity of Fungi of the Genus Aspergillus Section Nigri on Maize and Small Grains.Foods. 2025 Jun 19;14(12):2146. doi: 10.3390/foods14122146. Foods. 2025. PMID: 40565755 Free PMC article.
-
Genome-wide patterns of noncoding and protein-coding sequence variation in the major fungal pathogen Aspergillus fumigatus.G3 (Bethesda). 2024 Jul 8;14(7):jkae091. doi: 10.1093/g3journal/jkae091. G3 (Bethesda). 2024. PMID: 38696662 Free PMC article.
-
Population Genomics of Aspergillus sojae is Shaped by the Food Environment.Genome Biol Evol. 2025 Apr 3;17(4):evaf067. doi: 10.1093/gbe/evaf067. Genome Biol Evol. 2025. PMID: 40195023 Free PMC article.
References
-
- Houbraken J, de Vries RP, Samson RA. 2014. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species, p 199–249. In Advances in applied microbiology - PubMed
-
- Aime MC, Miller AN, Aoki T, Bensch K, Cai L, Crous PW, Hawksworth DL, Hyde KD, Kirk PM, Lücking R, May TW, Malosso E, Redhead SA, Rossman AY, Stadler M, Thines M, Yurkov AM, Zhang N, Schoch CL. 2021. How to publish a new fungal species, or name, version 3.0. IMA Fungus 12:11. doi:10.1186/s43008-021-00063-1 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

