Novel Alphaproteobacteria transcribe genes for nitric oxide transformation at high levels in a marine oxygen-deficient zone
- PMID: 38445905
- PMCID: PMC11022542
- DOI: 10.1128/aem.02099-23
Novel Alphaproteobacteria transcribe genes for nitric oxide transformation at high levels in a marine oxygen-deficient zone
Abstract
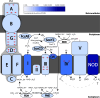
Marine oxygen-deficient zones (ODZs) are portions of the ocean where intense nitrogen loss occurs primarily via denitrification and anammox. Despite many decades of study, the identity of the microbes that catalyze nitrogen loss in ODZs is still being elucidated. Intriguingly, high transcription of genes in the same family as the nitric oxide dismutase (nod) gene from Methylomirabilota has been reported in the anoxic core of ODZs. Here, we show that the most abundantly transcribed nod genes in the Eastern Tropical North Pacific ODZ belong to a new order (UBA11136) of Alphaproteobacteria, rather than Methylomirabilota as previously assumed. Gammaproteobacteria and Planctomycetia also transcribe nod, but at lower relative abundance than UBA11136 in the upper ODZ. The nod-transcribing Alphaproteobacteria likely use formaldehyde and formate as a source of electrons for aerobic respiration, with additional electrons possibly from sulfide oxidation. They also transcribe multiheme cytochrome (here named ptd) genes for a putative porin-cytochrome protein complex of unknown function, potentially involved in extracellular electron transfer. Molecular oxygen for aerobic respiration may originate from nitric oxide dismutation via cryptic oxygen cycling. Our results implicate Alphaproteobacteria order UBA11136 as a significant player in marine nitrogen loss and highlight their potential in one-carbon, nitrogen, and sulfur metabolism in ODZs.IMPORTANCEIn marine oxygen-deficient zones (ODZs), microbes transform bioavailable nitrogen to gaseous nitrogen, with nitric oxide as a key intermediate. The Eastern Tropical North Pacific contains the world's largest ODZ, but the identity of the microbes transforming nitric oxide remains unknown. Here, we show that highly transcribed nitric oxide dismutase (nod) genes belong to Alphaproteobacteria of the novel order UBA11136, which lacks cultivated isolates. These Alphaproteobacteria show evidence for aerobic respiration, using oxygen potentially sourced from nitric oxide dismutase, and possess a novel porin-cytochrome protein complex with unknown function. Gammaproteobacteria and Planctomycetia transcribe nod at lower levels. Our results pinpoint the microbes mediating a key step in marine nitrogen loss and reveal an unexpected predicted metabolism for marine Alphaproteobacteria.
Keywords: Alphaproteobacteria; denitrification; marine; nitric oxide; nitrogen; oxygen; oxygen-deficient zone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Oxygen at nanomolar levels reversibly suppresses process rates and gene expression in anammox and denitrification in the oxygen minimum zone off northern Chile.mBio. 2014 Oct 28;5(6):e01966. doi: 10.1128/mBio.01966-14. mBio. 2014. PMID: 25352619 Free PMC article.
-
Uncultivated DPANN archaea are ubiquitous inhabitants of global oxygen-deficient zones with diverse metabolic potential.mBio. 2024 Mar 13;15(3):e0291823. doi: 10.1128/mbio.02918-23. Epub 2024 Feb 21. mBio. 2024. PMID: 38380943 Free PMC article.
-
Utilization of urea and cyanate in waters overlying and within the eastern tropical north Pacific oxygen deficient zone.FEMS Microbiol Ecol. 2018 Oct 1;94(10). doi: 10.1093/femsec/fiy138. FEMS Microbiol Ecol. 2018. PMID: 30016420
-
[Oxidation of inorganic sulfur compounds by obligatory organotrophic bacteria].Mikrobiologiia. 2003 Nov-Dec;72(6):725-39. Mikrobiologiia. 2003. PMID: 14768537 Review. Russian.
-
Cell biology and molecular basis of denitrification.Microbiol Mol Biol Rev. 1997 Dec;61(4):533-616. doi: 10.1128/mmbr.61.4.533-616.1997. Microbiol Mol Biol Rev. 1997. PMID: 9409151 Free PMC article. Review.
Cited by
-
Metabolic versatility of aerobic methane-oxidizing bacteria under anoxia in aquatic ecosystems.Environ Microbiol Rep. 2024 Oct;16(5):e70002. doi: 10.1111/1758-2229.70002. Environ Microbiol Rep. 2024. PMID: 39232853 Free PMC article. Review.
-
Widespread occurrence of dissolved oxygen anomalies, aerobic microbes, and oxygen-producing metabolic pathways in apparently anoxic environments.FEMS Microbiol Ecol. 2024 Oct 25;100(11):fiae132. doi: 10.1093/femsec/fiae132. FEMS Microbiol Ecol. 2024. PMID: 39327011 Free PMC article. Review.
-
Environment selected microbial function rather than taxonomic species in a plateau saline-alkaline wetland.Appl Environ Microbiol. 2025 Jul 23;91(7):e0220624. doi: 10.1128/aem.02206-24. Epub 2025 Jul 3. Appl Environ Microbiol. 2025. PMID: 40607849 Free PMC article.
References
-
- DeVries T, Deutsch C, Rafter PA, Primeau F. 2013. Marine denitrification rates determined from a global 3-D inverse model. Biogeosciences 10:2481–2496. doi: 10.5194/bg-10-2481-2013 - DOI
-
- Yang S, Chang BX, Warner MJ, Weber TS, Bourbonnais AM, Santoro AE, Kock A, Sonnerup RE, Bullister JL, Wilson ST, Bianchi D. 2020. Global reconstruction reduces the uncertainty of oceanic nitrous oxide emissions and reveals a vigorous seasonal cycle. Proc Natl Acad Sci U S A 117:11954–11960. doi: 10.1073/pnas.1921914117 - DOI - PMC - PubMed
-
- Frey C, Bange HW, Achterberg EP, Jayakumar A, Löscher CR, Arévalo-Martínez DL, León-Palmero E, Sun M, Sun X, Xie RC, Oleynik S, Ward BB. 2020. Regulation of nitrous oxide production in low-oxygen waters off the coast of Peru. Biogeosciences 17:2263–2287. doi: 10.5194/bg-17-2263-2020 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

