Identification and characterization of a fungal cutinase-like enzyme CpCut1 from Cladosporium sp. P7 for polyurethane degradation
- PMID: 38445906
- PMCID: PMC11022569
- DOI: 10.1128/aem.01477-23
Identification and characterization of a fungal cutinase-like enzyme CpCut1 from Cladosporium sp. P7 for polyurethane degradation
Abstract
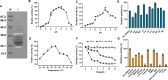
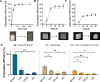
Plastic degradation by biological systems emerges as a prospective avenue for addressing the pressing global concern of plastic waste accumulation. The intricate chemical compositions and diverse structural facets inherent to polyurethanes (PU) substantially increase the complexity associated with PU waste management. Despite the extensive research endeavors spanning over decades, most known enzymes exhibit a propensity for hydrolyzing waterborne PU dispersion (i.e., the commercial Impranil DLN-SD), with only a limited capacity for the degradation of bulky PU materials. Here, we report a novel cutinase (CpCut1) derived from Cladosporium sp. P7, which demonstrates remarkable efficiency in the degrading of various polyester-PU materials. After 12-h incubation at 55°C, CpCut1 was capable of degrading 40.5% and 20.6% of thermoplastic PU film and post-consumer foam, respectively, while achieving complete depolymerization of Impranil DLN-SD. Further analysis of the degradation intermediates suggested that the activity of CpCut1 primarily targeted the ester bonds within the PU soft segments. The versatile performance of CpCut1 against a spectrum of polyester-PU materials positions it as a promising candidate for the bio-recycling of waste plastics.IMPORTANCEPolyurethane (PU) has a complex chemical composition that frequently incorporates a variety of additives, which poses significant obstacles to biodegradability and recyclability. Recent advances have unveiled microbial degradation and enzymatic depolymerization as promising waste PU disposal strategies. In this study, we identified a gene encoding a cutinase from the PU-degrading fungus Cladosporium sp. P7, which allowed the expression, purification, and characterization of the recombinant enzyme CpCut1. Furthermore, this study identified the products derived from the CpCut1 catalyzed PU degradation and proposed its underlying mechanism. These findings highlight the potential of this newly discovered fungal cutinase as a remarkably efficient tool in the degradation of PU materials.
Keywords: Cladosporium sp.; biodegradation; fungal cutinase; polymer recycling; polyurethane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Biodegradation of polyester polyurethane by Cladosporium sp. P7: Evaluating its degradation capacity and metabolic pathways.J Hazard Mater. 2023 Apr 15;448:130776. doi: 10.1016/j.jhazmat.2023.130776. Epub 2023 Jan 11. J Hazard Mater. 2023. PMID: 36706489
-
Biodegradation of polyester polyurethane by the marine fungus Cladosporium halotolerans 6UPA1.J Hazard Mater. 2022 Sep 5;437:129406. doi: 10.1016/j.jhazmat.2022.129406. Epub 2022 Jun 18. J Hazard Mater. 2022. PMID: 35753302
-
Novel polyurethane-degrading cutinase BaCut1 from Blastobotrys sp. G-9 with potential role in plastic bio-recycling.J Hazard Mater. 2024 Jul 5;472:134493. doi: 10.1016/j.jhazmat.2024.134493. Epub 2024 Apr 30. J Hazard Mater. 2024. PMID: 38696960
-
Solving the plastic dilemma: the fungal and bacterial biodegradability of polyurethanes.World J Microbiol Biotechnol. 2023 Mar 17;39(5):122. doi: 10.1007/s11274-023-03558-8. World J Microbiol Biotechnol. 2023. PMID: 36929307 Free PMC article. Review.
-
Microbial degradation of polyurethane, polyester polyurethanes and polyether polyurethanes.Appl Microbiol Biotechnol. 1999 Feb;51(2):134-40. doi: 10.1007/s002530051373. Appl Microbiol Biotechnol. 1999. PMID: 10091317 Review.
Cited by
-
Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.Adv Sci (Weinh). 2025 Apr;12(13):e2416019. doi: 10.1002/advs.202416019. Epub 2025 Feb 7. Adv Sci (Weinh). 2025. PMID: 39921299 Free PMC article.
-
PUR-GEN: A web server for automated generation of polyurethane fragment libraries.Comput Struct Biotechnol J. 2024 Dec 12;27:127-136. doi: 10.1016/j.csbj.2024.12.004. eCollection 2025. Comput Struct Biotechnol J. 2024. PMID: 39845943 Free PMC article.
-
Discovery of two novel cutinases from a gut yeast of plastic-eating mealworm for polyester depolymerization.Appl Environ Microbiol. 2025 Apr 23;91(4):e0256224. doi: 10.1128/aem.02562-24. Epub 2025 Apr 2. Appl Environ Microbiol. 2025. PMID: 40172219 Free PMC article.
References
-
- Plastics Europe . 2022. Plastics – the facts 2022 – an analysis of European plastics production, demand and waste data. Available from: https://plasticseurope.org
-
- Calvo-Correas T, Benitez M, Larraza I, Ugarte L, Peña-Rodríguez C, Eceiza A. 2022. Advanced and traditional processing of thermoplastic polyurethane waste. Polym Degrad Stab 198:109880. doi: 10.1016/j.polymdegradstab.2022.109880 - DOI
-
- Zia KM, Bhatti HN, Ahmad Bhatti I. 2007. Methods for polyurethane and polyurethane composites, recycling and recovery: a review. React Funct Polym 67:675–692. doi: 10.1016/j.reactfunctpolym.2007.05.004 - DOI
-
- Wang XK, Chen HB, Chen CG, Li HM. 2011. Chemical degradation of thermoplastic polyurethane for recycling polyether polyol. Fibers Polym 12:857–863. doi: 10.1007/s12221-011-0857-y - DOI
-
- Skleničková K, Abbrent S, Halecký M, Kočí V, Beneš H. 2022. Biodegradability and ecotoxicity of polyurethane foams: a review. Crit Rev Env Sci Tec 52:157–202. doi: 10.1080/10643389.2020.1818496 - DOI
Publication types
MeSH terms
Substances
Grants and funding
- 2021YFC2103600/MOST | National Key Research and Development Program of China (NKPs)
- 22278219/MOST | National Natural Science Foundation of China (NSFC)
- 31961133017/MOST | National Natural Science Foundation of China (NSFC)
- 32002316/MOST | National Natural Science Foundation of China (NSFC)
- BK20211591/Natural Science Foundation of Jiangsu Province of China for Excellent Young Scholars
LinkOut - more resources
Full Text Sources

