Novel influenza A viruses in pigs with zoonotic potential, Chile
- PMID: 38446039
- PMCID: PMC10986610
- DOI: 10.1128/spectrum.02181-23
Novel influenza A viruses in pigs with zoonotic potential, Chile
Abstract
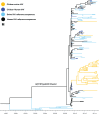
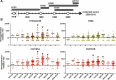
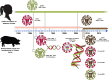
Novel H1N2 and H3N2 swine influenza A viruses (IAVs) have recently been identified in Chile. The objective of this study was to evaluate their zoonotic potential. We perform phylogenetic analyses to determine the genetic origin and evolution of these viruses, and a serological analysis to determine the level of cross-protective antibodies in the human population. Eight genotypes were identified, all with pandemic H1N1 2009-like internal genes. H1N1 and H1N2 were the subtypes more commonly detected. Swine H1N2 and H3N2 IAVs had hemagglutinin and neuraminidase lineages genetically divergent from IAVs reported worldwide, including human vaccine strains. These genes originated from human seasonal viruses were introduced into the swine population since the mid-1980s. Serological data indicate that the general population is susceptible to the H3N2 virus and that elderly and young children also lack protective antibodies against the H1N2 strains, suggesting that these viruses could be potential zoonotic threats. Continuous IAV surveillance and monitoring of the swine and human populations is strongly recommended.IMPORTANCEIn the global context, where swine serve as crucial intermediate hosts for influenza A viruses (IAVs), this study addresses the pressing concern of the zoonotic potential of novel reassortant strains. Conducted on a large scale in Chile, it presents a comprehensive account of swine influenza A virus diversity, covering 93.8% of the country's industrialized swine farms. The findings reveal eight distinct swine IAV genotypes, all carrying a complete internal gene cassette of pandemic H1N1 2009 origin, emphasizing potential increased replication and transmission fitness. Genetic divergence of H1N2 and H3N2 IAVs from globally reported strains raises alarms, with evidence suggesting introductions from human seasonal viruses since the mid-1980s. A detailed serological analysis underscores the zoonotic threat, indicating susceptibility in the general population to swine H3N2 and a lack of protective antibodies in vulnerable demographics. These data highlight the importance of continuous surveillance, providing crucial insights for global health organizations.
Keywords: influenza A virus; pigs; zoonosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Influenza A Viruses of Swine (IAV-S) in Vietnam from 2010 to 2015: Multiple Introductions of A(H1N1)pdm09 Viruses into the Pig Population and Diversifying Genetic Constellations of Enzootic IAV-S.J Virol. 2016 Dec 16;91(1):e01490-16. doi: 10.1128/JVI.01490-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795418 Free PMC article.
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Serological and molecular epidemiological study on swine influenza in Zambia.Transbound Emerg Dis. 2022 Jul;69(4):e931-e943. doi: 10.1111/tbed.14373. Epub 2021 Nov 16. Transbound Emerg Dis. 2022. PMID: 34724353
-
Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China.mBio. 2018 Jun 5;9(3):e00909-18. doi: 10.1128/mBio.00909-18. mBio. 2018. PMID: 29871917 Free PMC article.
-
Swine influenza viruses: an Asian perspective.Curr Top Microbiol Immunol. 2013;370:147-72. doi: 10.1007/82_2011_195. Curr Top Microbiol Immunol. 2013. PMID: 22266639 Review.
Cited by
-
A Computationally Optimized Broadly Reactive Hemagglutinin and Neuraminidase Vaccine Boosts Antibody-Secreting Cells and Induces a Robust Serological Response, Preventing Lung Damage in a Pre-Immune Model.Vaccines (Basel). 2024 Jun 24;12(7):706. doi: 10.3390/vaccines12070706. Vaccines (Basel). 2024. PMID: 39066344 Free PMC article.
-
Novel introductions of human-origin H3N2 influenza viruses in swine, Chile.Front Vet Sci. 2025 Jan 9;11:1505497. doi: 10.3389/fvets.2024.1505497. eCollection 2024. Front Vet Sci. 2025. PMID: 39850583 Free PMC article.
-
Circulation and Spillover of pdmH1N1 Influenza A Virus at an Educational Swine Farm in Chile, 2019-2023.Viruses. 2025 Apr 28;17(5):635. doi: 10.3390/v17050635. Viruses. 2025. PMID: 40431647 Free PMC article.
-
Cross-protection of commercial vaccines against Chilean swine influenza A virus using the guinea pig model as a surrogate.Front Vet Sci. 2023 Sep 20;10:1245278. doi: 10.3389/fvets.2023.1245278. eCollection 2023. Front Vet Sci. 2023. PMID: 37799404 Free PMC article.
References
-
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, Uyeki TM, Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team . 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360:2605–2615. doi:10.1056/NEJMoa0903810 - DOI - PubMed
-
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. 2014. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Hlth 61:4–17. doi:10.1111/zph.12049 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

