Directed evolution of the fluorescent protein CGP with in situ biosynthesized noncanonical amino acids
- PMID: 38446072
- PMCID: PMC11022568
- DOI: 10.1128/aem.01863-23
Directed evolution of the fluorescent protein CGP with in situ biosynthesized noncanonical amino acids
Abstract
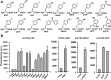
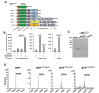
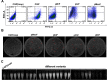
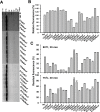
The incorporation of noncanonical amino acids (ncAAs) into proteins can enhance their function beyond the abilities of canonical amino acids and even generate new functions. However, the ncAAs used for such research are usually chemically synthesized, which is expensive and hinders their application on large industrial scales. We believe that the biosynthesis of ncAAs using metabolic engineering and their employment in situ in target protein engineering with genetic code expansion could overcome these limitations. As a proof of principle, we biosynthesized four ncAAs, O-L-methyltyrosine, 3,4-dihydroxy-L-phenylalanine, 5-hydroxytryptophan, and 5-chloro-L-tryptophan using metabolic engineering and directly evolved the fluorescent consensus green protein (CGP) by combination with nine other exogenous ncAAs in Escherichia coli. After screening a TAG scanning library expressing 13 ncAAs, several variants with enhanced fluorescence and stability were identified. The variants CGPV3pMeoF/K190pMeoF and CGPG20pMeoF/K190pMeoF expressed with biosynthetic O-L-methyltyrosine showed an approximately 1.4-fold improvement in fluorescence compared to the original level, and a 2.5-fold improvement in residual fluorescence after heat treatment. Our results demonstrated the feasibility of integrating metabolic engineering, genetic code expansion, and directed evolution in engineered cells to employ biosynthetic ncAAs in protein engineering. These results could further promote the application of ncAAs in protein engineering and enzyme evolution.
Importance: Noncanonical amino acids (ncAAs) have shown great potential in protein engineering and enzyme evolution through genetic code expansion. However, in most cases, ncAAs must be provided exogenously during protein expression, which hinders their application, especially when they are expensive or have poor cell membrane penetration. Engineering cells with artificial metabolic pathways to biosynthesize ncAAs and employing them in situ for protein engineering and enzyme evolution could facilitate their application and reduce costs. Here, we attempted to evolve the fluorescent consensus green protein (CGP) with biosynthesized ncAAs. Our results demonstrated the feasibility of using biosynthesized ncAAs in protein engineering, which could further stimulate the application of ncAAs in bioengineering and biomedicine.
Keywords: fluorescent protein; genetic code expansion; metabolic engineering; noncanonical amino acids; protein engineering.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Expanding the Structural Diversity of Protein Building Blocks with Noncanonical Amino Acids Biosynthesized from Aromatic Thiols.Angew Chem Int Ed Engl. 2021 Apr 26;60(18):10040-10048. doi: 10.1002/anie.202014540. Epub 2021 Mar 23. Angew Chem Int Ed Engl. 2021. PMID: 33570250
-
Expanding the eukaryotic genetic code with a biosynthesized 21st amino acid.Protein Sci. 2022 Oct;31(10):e4443. doi: 10.1002/pro.4443. Protein Sci. 2022. PMID: 36173166 Free PMC article.
-
Protein Expression with Biosynthesized Noncanonical Amino Acids.Methods Mol Biol. 2023;2676:87-100. doi: 10.1007/978-1-0716-3251-2_6. Methods Mol Biol. 2023. PMID: 37277626
-
Engineering enzymes for noncanonical amino acid synthesis.Chem Soc Rev. 2018 Dec 21;47(24):8980-8997. doi: 10.1039/c8cs00665b. Epub 2018 Oct 3. Chem Soc Rev. 2018. PMID: 30280154 Free PMC article. Review.
-
Advances in Biosynthesis of Non-Canonical Amino Acids (ncAAs) and the Methods of ncAAs Incorporation into Proteins.Molecules. 2023 Sep 21;28(18):6745. doi: 10.3390/molecules28186745. Molecules. 2023. PMID: 37764520 Free PMC article. Review.
Cited by
-
Directed evolution of L-DOPA sensing and production enables efficient incorporation of an expanded genetic code into proteins.bioRxiv [Preprint]. 2025 Jun 26:2025.06.26.661824. doi: 10.1101/2025.06.26.661824. bioRxiv. 2025. PMID: 40667249 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

