Carbapenemase-producing Enterobacterales strains causing infections in companion animals-Portugal
- PMID: 38446073
- PMCID: PMC10986603
- DOI: 10.1128/spectrum.03416-23
Carbapenemase-producing Enterobacterales strains causing infections in companion animals-Portugal
Abstract
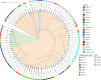
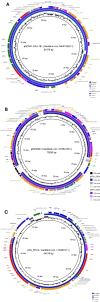
An increase in Klebsiella pneumoniae carbapenem-resistant human nosocomial strains is occurring in Europe, namely with the blaOXA-48-like and blaKPC-like genes. We determined the prevalence of carbapenemase-producing Enterobacterales clinical strains in companion animals in Portugal and characterized their mobile genetic elements. Susceptibility data of a consecutive collection of 977 Enterobacterales clinical strains from a Portuguese private veterinary diagnostic laboratory were evaluated (January-December 2020). Additional phenotypical and genotypical assays were performed in a subset of 261 strains with a resistant phenotype. Whole-genome sequencing was performed for carbapenemase-producing strains. The frequency of carbapenemase-producing Enterobacterales clinical strains in companion animals in Portugal was 0.51% (n = 5/977). Thus, five strains were characterized: (i) one OXA-181-producing K. pneumoniae ST273, (ii) two KPC-3-producing K. pneumoniae ST147; (iii) one KPC-3-producing K. pneumoniae ST392; and (iv) one OXA-48-producing E. coli ST127. The blaKPC-3 gene was located on transposon Tn4401d on IncFIA type plasmid for the K. pneumoniae ST147 strains and on a IncN-type plasmid for the K. pneumoniae ST392 strain, while blaOXA-181 gene was located on an IncX3 plasmid. All de novo assembled plasmids and plasmid-encoded transposons harboring carbapenemase genes were homologous to those previously described in the human healthcare. No plasmid replicons were detected on the OXA-48-producing E. coli ST127. The dissemination of carbapenem resistance is occurring horizontally via plasmid spreading from the human high burden carbapenem resistance setting to the companion animal sector. Furthermore, companion animals may act as reservoirs of carbapenem resistance. Implementation of carbapenemase detection methods in routine clinical veterinary microbiology is urgently needed.
Importance: This is the first study on the prevalence of carbapenemase-producing Enterobacterales (CPE) clinical strains from companion animals in Portugal. Despite the generally low prevalence of CPE in companion animals, it is imperative for veterinary diagnostic laboratories to employ diagnostic methods for carbapenemase detection. The resemblance found in the mobile genetic elements transporting carbapenemase genes between veterinary medicine and human medicine implies a potential circulation within a One Health framework.
Keywords: E. coli; KPC-3; Klebsiella pneumoniae; OXA-181; OXA-48; diagnostics; veterinary.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- WHO Regional Office for Europe/European Centre for Disease Prevention and Control . 2022. Antimicrobial resistance surveillance in Europe 2022 – 2020
-
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, EuSCAPE Working Group, ESGEM Study Group . 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi:10.1038/s41564-019-0492-8 - DOI - PMC - PubMed
-
- Mendes G, Ramalho JF, Bruschy-Fonseca A, Lito L, Duarte A, Melo-Cristino J, Caneiras C. 2022. Whole‐genome sequencing enables molecular characterization of non‐clonal group 258 high‐risk clones (ST13, ST17, ST147 and ST307) among carbapenem‐resistant Klebsiella pneumoniae from a tertiary university hospital centre in Portugal. Microorganisms 10:416. doi:10.3390/microorganisms10020416 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2020.06540.BD/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- 2020.07562.BD/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- UI/BD/153070/2022/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- UIDB/00276/2020/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- UID/04413/2020/MEC | Fundação para a Ciência e a Tecnologia (FCT)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

