Deep Learning-Augmented ECG Analysis for Screening and Genotype Prediction of Congenital Long QT Syndrome
- PMID: 38446445
- PMCID: PMC10918571
- DOI: 10.1001/jamacardio.2024.0039
Deep Learning-Augmented ECG Analysis for Screening and Genotype Prediction of Congenital Long QT Syndrome
Abstract
Importance: Congenital long QT syndrome (LQTS) is associated with syncope, ventricular arrhythmias, and sudden death. Half of patients with LQTS have a normal or borderline-normal QT interval despite LQTS often being detected by QT prolongation on resting electrocardiography (ECG).
Objective: To develop a deep learning-based neural network for identification of LQTS and differentiation of genotypes (LQTS1 and LQTS2) using 12-lead ECG.
Design, setting, and participants: This diagnostic accuracy study used ECGs from patients with suspected inherited arrhythmia enrolled in the Hearts in Rhythm Organization Registry (HiRO) from August 2012 to December 2021. The internal dataset was derived at 2 sites and an external validation dataset at 4 sites within the HiRO Registry; an additional cross-sectional validation dataset was from the Montreal Heart Institute. The cohort with LQTS included probands and relatives with pathogenic or likely pathogenic variants in KCNQ1 or KCNH2 genes with normal or prolonged corrected QT (QTc) intervals.
Exposures: Convolutional neural network (CNN) discrimination between LQTS1, LQTS2, and negative genetic test results.
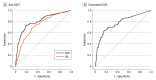
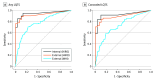
Main outcomes and measures: The main outcomes were area under the curve (AUC), F1 scores, and sensitivity for detecting LQTS and differentiating genotypes using a CNN method compared with QTc-based detection.
Results: A total of 4521 ECGs from 990 patients (mean [SD] age, 42 [18] years; 589 [59.5%] female) were analyzed. External validation within the national registry (101 patients) demonstrated the CNN's high diagnostic capacity for LQTS detection (AUC, 0.93; 95% CI, 0.89-0.96) and genotype differentiation (AUC, 0.91; 95% CI, 0.86-0.96). This surpassed expert-measured QTc intervals in detecting LQTS (F1 score, 0.84 [95% CI, 0.78-0.90] vs 0.22 [95% CI, 0.13-0.31]; sensitivity, 0.90 [95% CI, 0.86-0.94] vs 0.36 [95% CI, 0.23-0.47]), including in patients with normal or borderline QTc intervals (F1 score, 0.70 [95% CI, 0.40-1.00]; sensitivity, 0.78 [95% CI, 0.53-0.95]). In further validation in a cross-sectional cohort (406 patients) of high-risk patients and genotype-negative controls, the CNN detected LQTS with an AUC of 0.81 (95% CI, 0.80-0.85), which was better than QTc interval-based detection (AUC, 0.74; 95% CI, 0.69-0.78).
Conclusions and relevance: The deep learning model improved detection of congenital LQTS from resting ECGs and allowed for differentiation between the 2 most common genetic subtypes. Broader validation over an unselected general population may support application of this model to patients with suspected LQTS.
Conflict of interest statement
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

