Circulating hematopoietic stem/progenitor cell subsets contribute to human hematopoietic homeostasis
- PMID: 38446574
- PMCID: PMC11106755
- DOI: 10.1182/blood.2023022666
Circulating hematopoietic stem/progenitor cell subsets contribute to human hematopoietic homeostasis
Abstract
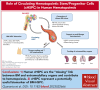
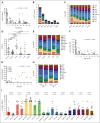
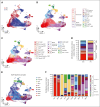
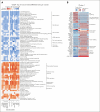
In physiological conditions, few circulating hematopoietic stem/progenitor cells (cHSPCs) are present in the peripheral blood, but their contribution to human hematopoiesis remain unsolved. By integrating advanced immunophenotyping, single-cell transcriptional and functional profiling, and integration site (IS) clonal tracking, we unveiled the biological properties and the transcriptional features of human cHSPC subpopulations in relationship to their bone marrow (BM) counterpart. We found that cHSPCs reduced in cell count over aging and are enriched for primitive, lymphoid, and erythroid subpopulations, showing preactivated transcriptional and functional state. Moreover, cHSPCs have low expression of multiple BM-retention molecules but maintain their homing potential after xenotransplantation. By generating a comprehensive human organ-resident HSPC data set based on single-cell RNA sequencing data, we detected organ-specific seeding properties of the distinct trafficking HSPC subpopulations. Notably, circulating multi-lymphoid progenitors are primed for seeding the thymus and actively contribute to T-cell production. Human clonal tracking data from patients receiving gene therapy (GT) also showed that cHSPCs connect distant BM niches and participate in steady-state hematopoietic production, with primitive cHSPCs having the highest recirculation capability to travel in and out of the BM. Finally, in case of hematopoietic impairment, cHSPCs composition reflects the BM-HSPC content and might represent a biomarker of the BM state for clinical and research purposes. Overall, our comprehensive work unveiled fundamental insights into the in vivo dynamics of human HSPC trafficking and its role in sustaining hematopoietic homeostasis. GT patients' clinical trials were registered at ClinicalTrials.gov (NCT01515462 and NCT03837483) and EudraCT (2009-017346-32 and 2018-003842-18).
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




Comment in
-
Patrolling progenitors: a first responder team.Blood. 2024 May 9;143(19):1883-1884. doi: 10.1182/blood.2024024443. Blood. 2024. PMID: 38722657 No abstract available.
References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

