The RNA m6A reader IGF2BP3 regulates NFAT1/IRF1 axis-mediated anti-tumor activity in gastric cancer
- PMID: 38448411
- PMCID: PMC10917814
- DOI: 10.1038/s41419-024-06566-0
The RNA m6A reader IGF2BP3 regulates NFAT1/IRF1 axis-mediated anti-tumor activity in gastric cancer
Abstract
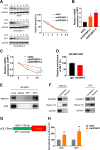
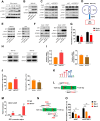
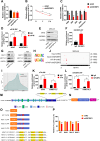
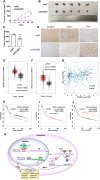
N6-methyladenosine (m6A) and its associated reader protein insulin like growth factor 2 mRNA binding protein 3 (IGF2BP3) are involved in tumor initiation and progression via regulating RNA metabolism. This study aims to investigate the biological function and clinical significance of IGF2BP3 in gastric cancer (GC). The clinical significance of IGF2BP3 was evaluated using tumor related databases and clinical tissues. The biological role and molecular mechanism of IGF2BP3 in GC progression were investigated by multi-omics analysis including Ribosome sequence (Ribo-seq), RNA sequence (RNA-seq) and m6A sequence (m6A-seq) combined with gain- and loss- of function experiments. IGF2BP3 expression is significantly elevated in GC tissues and associated with poor prognosis of GC patients. Knockdown of IGF2BP3 significantly weakens the migration and clonogenic ability, promotes the apoptosis, inhibits translation, and suppresses in vitro growth and progression of GC cells. Mechanistically, IGF2BP3 regulates the mRNA stability and translation of the nuclear factor of activated T cells 1(NFAT1) in a m6A dependent manner. Then NFAT1 induced by IGF2BP3 acts as a transcription factor (TF) to negatively regulates the promoter activities of interferon regulatory factor 1 (IRF1) to inhibit its expression. Inhibition of IGF2BP3-induced expression of IRF1 activates interferon (IFN) signaling pathway and then exerts its anti-tumor effect. Elevated IGF2BP3 promotes in vivo and in vitro GC progression via regulation of NFAT1/IRF1 pathways. Targeted inhibition of IGF2BP3 might be a potential therapeutic approach for GC treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
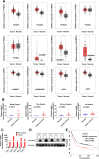
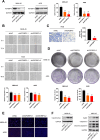
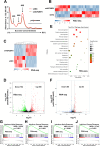
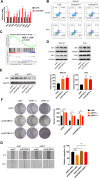
References
Publication types
MeSH terms
Substances
Grants and funding
- 81871702/National Natural Science Foundation of China (National Science Foundation of China)
- 82102494/National Natural Science Foundation of China (National Science Foundation of China)
- 81973343/National Natural Science Foundation of China (National Science Foundation of China)
- 2021M703706/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

