SARS-CoV-2 virulence factor ORF3a blocks lysosome function by modulating TBC1D5-dependent Rab7 GTPase cycle
- PMID: 38448435
- PMCID: PMC10918171
- DOI: 10.1038/s41467-024-46417-2
SARS-CoV-2 virulence factor ORF3a blocks lysosome function by modulating TBC1D5-dependent Rab7 GTPase cycle
Abstract
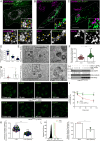
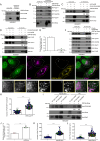
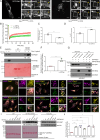
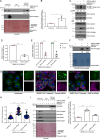
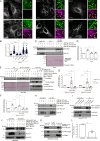
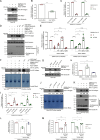
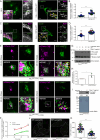
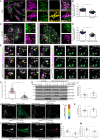
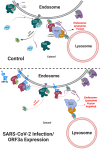
SARS-CoV-2, the causative agent of COVID-19, uses the host endolysosomal system for entry, replication, and egress. Previous studies have shown that the SARS-CoV-2 virulence factor ORF3a interacts with the lysosomal tethering factor HOPS complex and blocks HOPS-mediated late endosome and autophagosome fusion with lysosomes. Here, we report that SARS-CoV-2 infection leads to hyperactivation of the late endosomal and lysosomal small GTP-binding protein Rab7, which is dependent on ORF3a expression. We also observed Rab7 hyperactivation in naturally occurring ORF3a variants encoded by distinct SARS-CoV-2 variants. We found that ORF3a, in complex with Vps39, sequesters the Rab7 GAP TBC1D5 and displaces Rab7 from this complex. Thus, ORF3a disrupts the GTP hydrolysis cycle of Rab7, which is beneficial for viral production, whereas the Rab7 GDP-locked mutant strongly reduces viral replication. Hyperactivation of Rab7 in ORF3a-expressing cells impaired CI-M6PR retrieval from late endosomes to the trans-Golgi network, disrupting the biosynthetic transport of newly synthesized hydrolases to lysosomes. Furthermore, the tethering of the Rab7- and Arl8b-positive compartments was strikingly reduced upon ORF3a expression. As SARS-CoV-2 egress requires Arl8b, these findings suggest that ORF3a-mediated hyperactivation of Rab7 serves a multitude of functions, including blocking endolysosome formation, interrupting the transport of lysosomal hydrolases, and promoting viral egress.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Regulation of autophagy by SARS-CoV-2: The multifunctional contributions of ORF3a.J Med Virol. 2023 Jul;95(7):e28959. doi: 10.1002/jmv.28959. J Med Virol. 2023. PMID: 37485696 Review.
-
The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.Elife. 2023 Jan 25;12:e84477. doi: 10.7554/eLife.84477. Elife. 2023. PMID: 36695574 Free PMC article.
-
ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation.Dev Cell. 2021 Feb 22;56(4):427-442.e5. doi: 10.1016/j.devcel.2020.12.010. Epub 2020 Dec 16. Dev Cell. 2021. PMID: 33422265 Free PMC article.
-
The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.bioRxiv [Preprint]. 2022 Sep 3:2022.09.02.506428. doi: 10.1101/2022.09.02.506428. bioRxiv. 2022. Update in: Elife. 2023 Jan 25;12:e84477. doi: 10.7554/eLife.84477. PMID: 36263072 Free PMC article. Updated. Preprint.
-
Understanding the Role of SARS-CoV-2 ORF3a in Viral Pathogenesis and COVID-19.Front Microbiol. 2022 Mar 9;13:854567. doi: 10.3389/fmicb.2022.854567. eCollection 2022. Front Microbiol. 2022. PMID: 35356515 Free PMC article. Review.
Cited by
-
SARS-CoV-2 remodels the Golgi apparatus to facilitate viral assembly and secretion.PLoS Pathog. 2025 Jun 20;21(6):e1013295. doi: 10.1371/journal.ppat.1013295. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40540531 Free PMC article.
-
Impact of SARS-CoV-2 Wuhan and Omicron Variant Proteins on Type I Interferon Response.Viruses. 2025 Apr 15;17(4):569. doi: 10.3390/v17040569. Viruses. 2025. PMID: 40285011 Free PMC article.
-
Itaconate facilitates viral infection via alkylating GDI2 and retaining Rab GTPase on the membrane.Signal Transduct Target Ther. 2024 Dec 27;9(1):371. doi: 10.1038/s41392-024-02077-8. Signal Transduct Target Ther. 2024. PMID: 39730330 Free PMC article.
-
SARS-CoV-2 ORF3a accessory protein is a water-permeable channel that induces lysosome swelling.Commun Biol. 2025 Feb 4;8(1):170. doi: 10.1038/s42003-024-07442-5. Commun Biol. 2025. PMID: 39905220 Free PMC article.
-
SARS-CoV-2 encoded ORF3a interacts with YY1 to promote latent HCMV reactivation.PLoS Pathog. 2025 Jul 16;21(7):e1013344. doi: 10.1371/journal.ppat.1013344. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40668819 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

