Genome-wide characterization of circulating metabolic biomarkers
- PMID: 38448586
- PMCID: PMC10990933
- DOI: 10.1038/s41586-024-07148-y
Genome-wide characterization of circulating metabolic biomarkers
Abstract
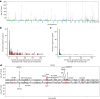
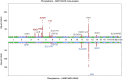
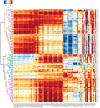
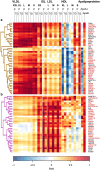
Genome-wide association analyses using high-throughput metabolomics platforms have led to novel insights into the biology of human metabolism1-7. This detailed knowledge of the genetic determinants of systemic metabolism has been pivotal for uncovering how genetic pathways influence biological mechanisms and complex diseases8-11. Here we present a genome-wide association study for 233 circulating metabolic traits quantified by nuclear magnetic resonance spectroscopy in up to 136,016 participants from 33 cohorts. We identify more than 400 independent loci and assign probable causal genes at two-thirds of these using manual curation of plausible biological candidates. We highlight the importance of sample and participant characteristics that can have significant effects on genetic associations. We use detailed metabolic profiling of lipoprotein- and lipid-associated variants to better characterize how known lipid loci and novel loci affect lipoprotein metabolism at a granular level. We demonstrate the translational utility of comprehensively phenotyped molecular data, characterizing the metabolic associations of intrahepatic cholestasis of pregnancy. Finally, we observe substantial genetic pleiotropy for multiple metabolic pathways and illustrate the importance of careful instrument selection in Mendelian randomization analysis, revealing a putative causal relationship between acetone and hypertension. Our publicly available results provide a foundational resource for the community to examine the role of metabolism across diverse diseases.
© 2024. The Author(s).
Conflict of interest statement
During the course of the project P.S. became a full-time employee of GlaxoSmithKline. V.S. has received an honorarium from Sanofi for consulting. V.S. also has an ongoing research collaboration with Bayer (outside the present study). A.M. is an employee of Genentech and a holder of Roche stock. N.v.Z. is currently employed by AstraZeneca PLC and is a shareholder in AstraZeneca. R.L.-G. is a part-time contractor of Metabolon Inc. During the course of the project J.Z. became a full-time employee of Novartis. A.I.d.H. is currently an employee of AbbVie. C.M. is funded by the Chronic Disease Research Foundation (CDRF). T.D.S. is co-founder and shareholder of ZOE Ltd. M.I.M. is an employee of Genentech and a holder of Roche stock. J.D. serves on scientific advisory boards for AstraZeneca, Novartis and UK Biobank, and has received multiple grants from academic, charitable and industry sources outside of the submitted work. A.S.B. reports institutional grants outside of this work from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Novartis, Regeneron and Sanofi. The other authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- MC_UU_00007/10/MRC_/Medical Research Council/United Kingdom
- 19169/CRUK_/Cancer Research UK/United Kingdom
- R01 DK062370/DK/NIDDK NIH HHS/United States
- MC_PC_15018/MRC_/Medical Research Council/United Kingdom
- 202802/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_17223/MRC_/Medical Research Council/United Kingdom
- G9815508/MRC_/Medical Research Council/United Kingdom
- CH/1996001/9454/BHF_/British Heart Foundation/United Kingdom
- MC_PC_19009/MRC_/Medical Research Council/United Kingdom
- P30 DK020572/DK/NIDDK NIH HHS/United States
- MC_PC_18026/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

