The hyphal-specific toxin candidalysin promotes fungal gut commensalism
- PMID: 38448595
- PMCID: PMC11230112
- DOI: 10.1038/s41586-024-07142-4
The hyphal-specific toxin candidalysin promotes fungal gut commensalism
Abstract
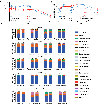
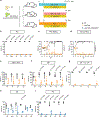
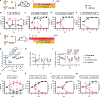
The fungus Candida albicans frequently colonizes the human gastrointestinal tract, from which it can disseminate to cause systemic disease. This polymorphic species can transition between growing as single-celled yeast and as multicellular hyphae to adapt to its environment. The current dogma of C. albicans commensalism is that the yeast form is optimal for gut colonization, whereas hyphal cells are detrimental to colonization but critical for virulence1-3. Here, we reveal that this paradigm does not apply to multi-kingdom communities in which a complex interplay between fungal morphology and bacteria dictates C. albicans fitness. Thus, whereas yeast-locked cells outcompete wild-type cells when gut bacteria are absent or depleted by antibiotics, hyphae-competent wild-type cells outcompete yeast-locked cells in hosts with replete bacterial populations. This increased fitness of wild-type cells involves the production of hyphal-specific factors including the toxin candidalysin4,5, which promotes the establishment of colonization. At later time points, adaptive immunity is engaged, and intestinal immunoglobulin A preferentially selects against hyphal cells1,6. Hyphal morphotypes are thus under both positive and negative selective pressures in the gut. Our study further shows that candidalysin has a direct inhibitory effect on bacterial species, including limiting their metabolic output. We therefore propose that C. albicans has evolved hyphal-specific factors, including candidalysin, to better compete with bacterial species in the intestinal niche.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
No competing interests noted.
Figures





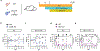




References
Additional references
-
- Guthrie C & Fink GR Guide to Yeast Genetics and Molecular Biology. (Academic Press, 1991).
-
- Liu H, Kohler J & Fink GR Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266, 1723–1726 (1994). - PubMed
-
- Reuss O, Vik A, Kolter R & Morschhauser J The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341, 119–127 (2004). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

