Lupus autoantibodies initiate neuroinflammation sustained by continuous HMGB1:RAGE signaling and reversed by increased LAIR-1 expression
- PMID: 38448779
- PMCID: PMC11141703
- DOI: 10.1038/s41590-024-01772-6
Lupus autoantibodies initiate neuroinflammation sustained by continuous HMGB1:RAGE signaling and reversed by increased LAIR-1 expression
Abstract
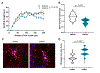
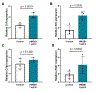
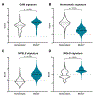
Cognitive impairment is a frequent manifestation of neuropsychiatric systemic lupus erythematosus, present in up to 80% of patients and leading to a diminished quality of life. In the present study, we used a model of lupus-like cognitive impairment that is initiated when antibodies that crossreact with excitatory neuronal receptors penetrate the hippocampus, causing immediate, self-limited, excitotoxic death of hippocampal neurons, which is then followed by a significant loss of dendritic complexity in surviving neurons. This injury creates a maladaptive equilibrium that is sustained in mice for at least 1 year. We identified a feedforward loop of microglial activation and microglia-dependent synapse elimination dependent on neuronal secretion of high mobility group box 1 protein (HMGB1) which binds the receptor for advanced glycation end products (RAGE) and leads to microglial secretion of C1q, upregulation of interleukin-10 with consequent downregulation of leukocyte-associated immunoglobulin-like receptor 1 (LAIR-1), an inhibitory receptor for C1q. Treatment with a centrally acting angiotensin-converting enzyme inhibitor or with an angiotensin-receptor blocker restored a healthy equilibrium, microglial quiescence and intact spatial memory.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures
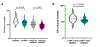






Update of
-
Lupus autoantibodies initiate a maladaptive equilibrium sustained by HMGB1:RAGE signaling and reversed by LAIR-1:C1q signaling.Res Sq [Preprint]. 2023 May 22:rs.3.rs-2870168. doi: 10.21203/rs.3.rs-2870168/v1. Res Sq. 2023. Update in: Nat Immunol. 2024 Apr;25(4):671-681. doi: 10.1038/s41590-024-01772-6. PMID: 37292843 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

