Correlation between large rearrangements and patient phenotypes in NF1 deletion syndrome: an update and review
- PMID: 38448973
- PMCID: PMC10919053
- DOI: 10.1186/s12920-024-01843-5
Correlation between large rearrangements and patient phenotypes in NF1 deletion syndrome: an update and review
Abstract
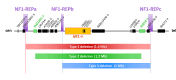
About 5-10% of neurofibromatosis type 1 (NF1) patients exhibit large genomic germline deletions that remove the NF1 gene and its flanking regions. The most frequent NF1 large deletion is 1.4 Mb, resulting from homologous recombination between two low copy repeats. This "type-1" deletion is associated with a severe clinical phenotype in NF1 patients, with several phenotypic manifestations including learning disability, a much earlier development of cutaneous neurofibromas, an increased tumour risk, and cardiovascular malformations. NF1 adjacent co-deleted genes could act as modifier loci for the specific clinical manifestations observed in deleted NF1 patients. Furthermore, other genetic modifiers (such as CNVs) not located at the NF1 locus could also modulate the phenotype observed in patients with large deletions. In this study, we analysed 22 NF1 deletion patients by genome-wide array-CGH with the aim (1) to correlate deletion length to observed phenotypic features and their severity in NF1 deletion syndrome, and (2) to identify whether the deletion phenotype could also be modulated by copy number variations elsewhere in the genome. We then review the role of co-deleted genes in the 1.4 Mb interval of type-1 deletions, and their possible implication in the main clinical features observed in this high-risk group of NF1 patients.
Keywords: CNV; Deletion; Genotype-phenotype correlation; NF1; Neurofibromatosis type 1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Vogt J, Bengesser K, Claes KBM, Wimmer K, Mautner V-F, van Minkelen R, et al. SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints. Genome Biol. 2014;15:R80. doi: 10.1186/gb-2014-15-6-r80. - DOI - PMC - PubMed
-
- Bengesser K, Vogt J, Mussotter T, Mautner V-F, Messiaen L, Cooper DN, et al. Analysis of crossover breakpoints yields new insights into the nature of the gene conversion events associated with large NF1 deletions mediated by nonallelic homologous recombination. Hum Mutat. 2014;35:215–26. doi: 10.1002/humu.22473. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

