Modification of Huntington's disease by short tandem repeats
- PMID: 38449714
- PMCID: PMC10917446
- DOI: 10.1093/braincomms/fcae016
Modification of Huntington's disease by short tandem repeats
Abstract
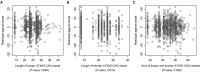
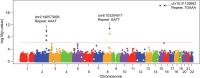
Expansions of glutamine-coding CAG trinucleotide repeats cause a number of neurodegenerative diseases, including Huntington's disease and several of spinocerebellar ataxias. In general, age-at-onset of the polyglutamine diseases is inversely correlated with the size of the respective inherited expanded CAG repeat. Expanded CAG repeats are also somatically unstable in certain tissues, and age-at-onset of Huntington's disease corrected for individual HTT CAG repeat length (i.e. residual age-at-onset), is modified by repeat instability-related DNA maintenance/repair genes as demonstrated by recent genome-wide association studies. Modification of one polyglutamine disease (e.g. Huntington's disease) by the repeat length of another (e.g. ATXN3, CAG expansions in which cause spinocerebellar ataxia 3) has also been hypothesized. Consequently, we determined whether age-at-onset in Huntington's disease is modified by the CAG repeats of other polyglutamine disease genes. We found that the CAG measured repeat sizes of other polyglutamine disease genes that were polymorphic in Huntington's disease participants but did not influence Huntington's disease age-at-onset. Additional analysis focusing specifically on ATXN3 in a larger sample set (n = 1388) confirmed the lack of association between Huntington's disease residual age-at-onset and ATXN3 CAG repeat length. Additionally, neither our Huntington's disease onset modifier genome-wide association studies single nucleotide polymorphism data nor imputed short tandem repeat data supported the involvement of other polyglutamine disease genes in modifying Huntington's disease. By contrast, our genome-wide association studies based on imputed short tandem repeats revealed significant modification signals for other genomic regions. Together, our short tandem repeat genome-wide association studies show that modification of Huntington's disease is associated with short tandem repeats that do not involve other polyglutamine disease-causing genes, refining the landscape of Huntington's disease modification and highlighting the importance of rigorous data analysis, especially in genetic studies testing candidate modifiers.
Keywords: ATXN3; Huntington’s disease; genetic modification; polyglutamine disease; short tandem repeat.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
J.F.G. was a Scientific Advisory Board member and had a financial interest in Triplet Therapeutics, Inc. His NIH-funded project is using genetic and genomic approaches to uncover other genes that significantly influence when diagnosable symptoms emerge and how rapidly they worsen in Huntington disease. The company is developing new therapeutic approaches to address triplet repeat disorders such Huntington's disease, myotonic dystrophy and SCAs. His interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. J.F.G. has also been a consultant for Wave Life Sciences USA, Inc., Biogen, Inc. and Pfizer, Inc. Within the last five years D.G.M. has been a scientific consultant and/or received an honoraria/stock options from AMO Pharma, Dyne, F. Hoffman-La Roche, LoQus23, Novartis, Ono Pharmaceuticals, Rgenta Therapeutics, Sanofi, Sarepta Therapeutics Inc, Script Biosciences, Triplet Therapeutics, and Vertex Pharmaceuticals and held research contracts with AMO Pharma and Vertex Pharmaceuticals. J.D.L. is a paid Advisory Board member for F. Hoffmann-La Roche Ltd and uniQure biopharma B.V., and he is a paid consultant for Vaccinex Inc, Wave Life Sciences USA Inc, Genentech Inc, Triplet Inc, and PTC Therapeutics Inc. T.H.M. is an associate member of the scientific advisory board of LoQus23 Therapeutics. L.J. was a member of the scientific advisory boards of LoQus23 Therapeutics and Triplet Therapeutics. V.C.W. was a Scientific Advisory Board member of Triplet Therapeutics, Inc., a company developing new therapeutic approaches to address triplet repeat disorders such Huntington's disease and myotonic dystrophy. Her financial interests in Triplet Therapeutics were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict of interest policies. She is a scientific advisory board member of LoQus23 Therapeutics and has provided paid consulting services to Alnylam, Acadia Pharmaceuticals Inc., Alnylam Inc., Biogen Inc. and Passage Bio. J.M.L consults for Life Edit Therapeutics and serves in the scientific advisory board of GenEdit, Inc.
Figures

References
-
- Di Prospero NA, Fischbeck KH. Therapeutics development for triplet repeat expansion diseases. Nat Rev Genet. 2005;6(10):756–765. - PubMed
-
- Ross CA. Polyglutamine pathogenesis: Emergence of unifying mechanisms for huntington's disease and related disorders. Neuron. 2002;35(5):819–822. - PubMed
-
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann Rev Neurosci. 2007;30:575–621. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
