Targeting Wnt signaling for improved glioma immunotherapy
- PMID: 38449858
- PMCID: PMC10915090
- DOI: 10.3389/fimmu.2024.1342625
Targeting Wnt signaling for improved glioma immunotherapy
Abstract
Introduction: Despite aggressive standard-of-care therapy, including surgery, radiation, and chemotherapy, glioblastoma recurrence is almost inevitable and uniformly lethal. Activation of glioma-intrinsic Wnt/β-catenin signaling is associated with a poor prognosis and the proliferation of glioma stem-like cells, leading to malignant transformation and tumor progression. Impressive results in a subset of cancers have been obtained using immunotherapies including anti-CTLA4, anti-PD-1, and anti-PD-L1 or chimeric antigen receptor (CAR) T cell therapies. However, the heterogeneity of tumors, low mutational burden, single antigen targeting, and associated antigen escape contribute to non-responsiveness and potential tumor recurrence despite these therapeutic efforts. In the current study, we determined the effects of the small molecule, highly specific Wnt/CBP (CREB Binding Protein)/β-catenin antagonist ICG-001, on glioma tumor cells and the tumor microenvironment (TME)-including its effect on immune cell infiltration, blood vessel decompression, and metabolic changes.
Methods: Using multiple glioma patient-derived xenografts cell lines and murine tumors (GL261, K-Luc), we demonstrated in vitro cytostatic effects and a switch from proliferation to differentiation after treatment with ICG-001.
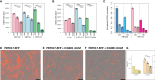
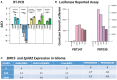
Results: In these glioma cell lines, we further demonstrated that ICG-001 downregulated the CBP/β-catenin target gene Survivin/BIRC5-a hallmark of Wnt/CBP/β-catenin inhibition. We found that in a syngeneic mouse model of glioma (K-luc), ICG-001 treatment enhanced tumor infiltration by CD3+ and CD8+ cells with increased expression of the vascular endothelial marker CD31 (PECAM-1). We also observed differential gene expression and induced immune cell infiltration in tumors pretreated with ICG-001 and then treated with CAR T cells as compared with single treatment groups or when ICG-001 treatment was administered after CAR T cell therapy.
Discussion: We conclude that specific Wnt/CBP/β-catenin antagonism results in pleotropic changes in the glioma TME, including glioma stem cell differentiation, modulation of the stroma, and immune cell activation and recruitment, thereby suggesting a possible role for enhancing immunotherapy in glioma patients.
Keywords: ICG-001; NanoString gene expression; Wnt signaling, pathway; differentiation; glioma; immunotherapy; proteomics.
Copyright © 2024 Gutova, Hibbard, Ma, Natri, Adhikarla, Chimge, Qiu, Nguyen, Melendez, Aguilar, Starr, Yin, Rockne, Ono, Banovich, Yuan, Brown and Kahn.
Conflict of interest statement
MK is a cofounder and holds equity in 3 + 2 Pharmaceuticals. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Sharma P, Siddiqui BA, Anandhan S, Yadav SS, Subudhi SK, Gao J, et al. . The next decade of immune checkpoint therapy. Cancer Discovery (2021) 11(4):838–57. doi: 10.1158/2159-8290.CD-20-1680 - DOI - PubMed
-
- Kahlert UD, Maciaczyk D, Doostkam S, Orr BA, Simons B, Bogiel T, et al. . Activation of canonical WNT/beta-catenin signaling enhances in vitro motility of glioblastoma cells by activation of ZEB1 and other activators of epithelial-to-mesenchymal transition. Cancer Lett (2012) 325(1):42–53. doi: 10.1016/j.canlet.2012.05.024 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

