Cell-free DNA from nail clippings as source of normal control for genomic studies in hematologic malignancies
- PMID: 38450530
- PMCID: PMC11443392
- DOI: 10.3324/haematol.2024.285054
Cell-free DNA from nail clippings as source of normal control for genomic studies in hematologic malignancies
Abstract
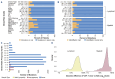
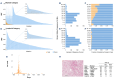
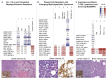
Comprehensive genomic sequencing is becoming a critical component in the assessment of hematologic malignancies, with broad implications for patients' management. In this context, unequivocally discriminating somatic from germline events is challenging but greatly facilitated by matched analysis of tumor:normal pairs of samples. In contrast to solid tumors, in hematologic malignancies conventional sources of normal control material (peripheral blood, buccal swabs, saliva) could be highly involved by the neoplastic process, rendering them unsuitable. In this work we describe our real-world experience using cell-free DNA (cfDNA) isolated from nail clippings as an alternate source of normal control material, through the dedicated review of 2,610 tumor:nail pairs comprehensively sequenced by MSK-IMPACT-heme. Overall, we found that nail cfDNA is a robust germline control for paired genomic studies. In a subset of patients, nail DNA may be contaminated by tumor DNA, reflecting unique attributes of the hematologic disease and transplant history. Contamination is generally low level, but significantly more common among patients with myeloid neoplasms (20.5%; 304/1,482) than among those with lymphoid diseases (5.4%; 61/1,128) and particularly enriched in myeloproliferative neoplasms with marked myelofibrosis. When identified in patients with lymphoid and plasma-cell neoplasms, mutations commonly reflected a myeloid profile and correlated with a concurrent/evolving clonal myeloid neoplasm. Donor DNA was identified in 22% (11/50) of nails collected after allogeneic stem-cell transplantation. In this cohort, an association with a recent history of graft-versus-host disease was identified. These findings should be considered as a potential limitation to the use of nails as a source of normal control DNA but could also provide important diagnostic information regarding the disease process.
Figures


References
-
- Kontopoulos I, Presslee S, Penkman K, Collins M. Preparation of bone powder for FTIR-ATR analysis: the particle size effect. Vibr Spectrosc. 2018;99:167-177.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

